Current location: Home > PRODUCTS > Next Generation Sequencing Series Products
PRODUCTS
(For Research Use Only)
Uterine leiomyomas are benign mesenchymal cell tumors of smooth muscle origin, displaying various morphologies, and are the most common benign tumors in women. The incidence in reproductive-aged women can reach up to 25%, and in perimenopausal women, it can be as high as 70% [1]. Most of these lesions are caused by MED12 mutations, with less common pathways involving FH biallelic gene inactivation, as well as chromosomal fragmentation and gene rearrangements [2]. MED12 gene mutations occur in over 70% of uterine leiomyoma patients, with the majority happening in exon 2 [3], and mutations in codon 44 account for more than half (36% to 96%) [4].
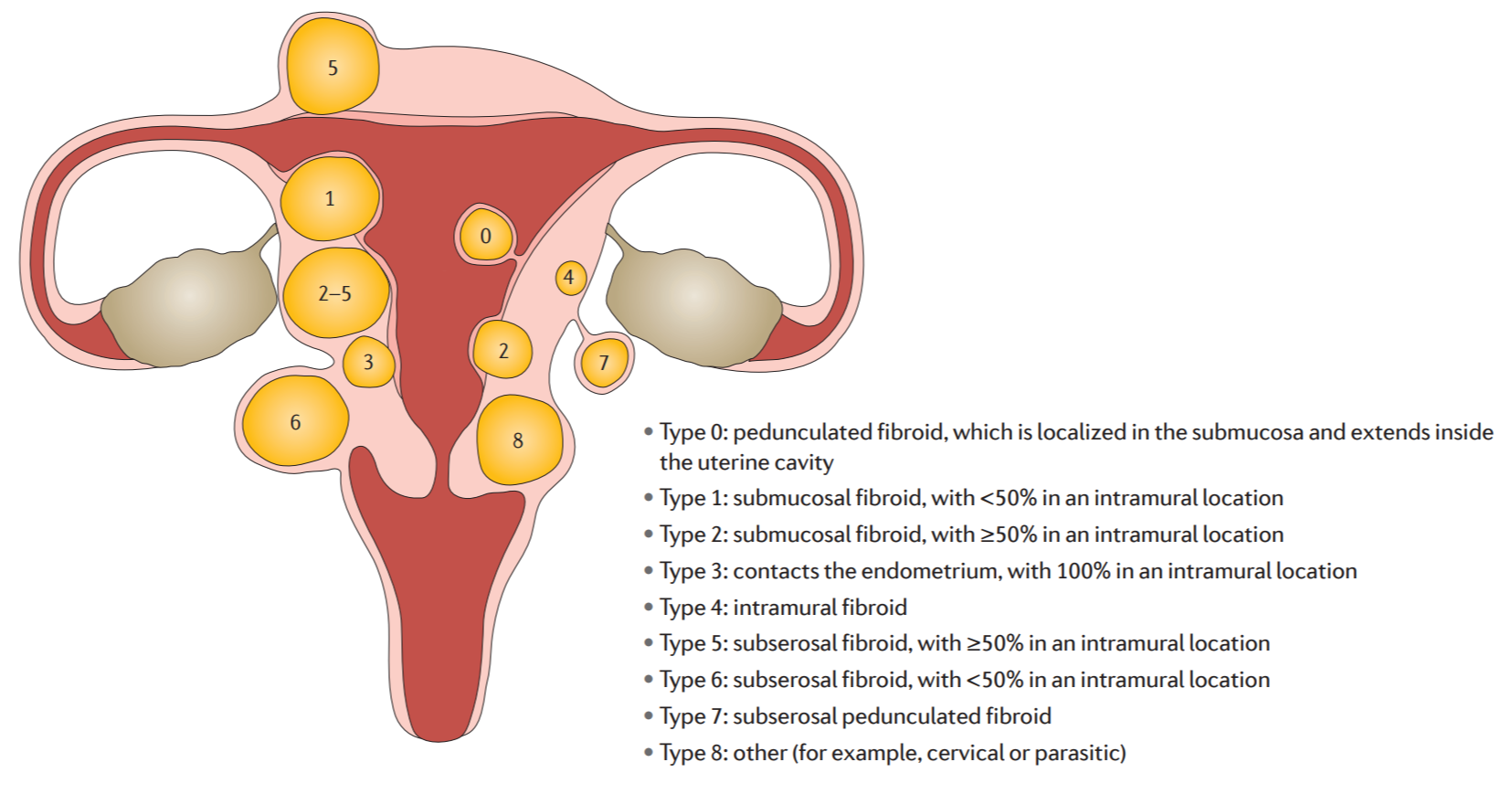
Federation International of Gynecology and Obstetrics (FIGO) uterine leiomyoma subclassification system [5]
HLRCC
FH-deficient type uterine leiomyomas are a rare subtype of uterine leiomyomas, accounting for approximately 0.4% to 1.6% of all uterine leiomyomas. They are associated with germline or somatic mutations in the FH gene. Hereditary leiomyomatosis and renal cell cancer (HLRCC) syndrome are caused by germline mutations in the FH gene, and it is an autosomal dominant inherited disorder characterized by multiple cutaneous leiomyomas, early-onset uterine leiomyomas, and renal cell carcinoma [6]. Since uterine leiomyomas are typically diagnosed early, easily detected, and have a favorable prognosis, they can serve as benign precursor tumors for HLRCC-related kidney cancer.
Relying solely on FH immunohistochemistry can result in the underdiagnosis of some patients, and genetic testing is considered the gold standard for diagnosing FH-deficient type uterine leiomyomas [6]. The FH gene has a total length of approximately 22 kb and is composed of 10 exons. Mutations in this gene exhibit a relatively high degree of randomness, with no specific high-frequency mutation hotspots. The most common mutation type is missense mutations (50% to 60%), followed by nonsense mutations and frameshift mutations (together accounting for approximately 30%) [7]. Copy number variations (CNV) are also one of the molecular genetic mechanisms of FH mutations [6]. Some cases of FH-deficient uterine leiomyomas and FH-deficient renal cell carcinoma result from somatic mutations in the FH gene [6-7], so suspected patients should undergo full exon testing for both germline and somatic FH mutations [7].
Based on the phase II clinical trial ( NCT01130519 ), the objective response rate ( ORR ) of HLRCC patients treated with bevacizumab combined with erlotinib was 64 %, and the ORR of patients with sporadic papillary renal cell carcinoma ( pRCC ) was 37 %. The median progression-free survival ( mPFS ) of HLRCC patients was 21.1 months, and the mPFS of sporadic pRCC patients was 8.7 months. The NCCN guidelines recommend the use of bevacizumab combined with erlotinib in patients with advanced pRCC ( including HLRCC ) (category 2A).
DETECTION CONTENT
Qualitatively detect the complete coding region (CDS) of the FH gene and the MED12 gene hotspot region in tissue and/or blood samples to assist in the diagnosis of HLRCC.
1、Auxiliary diagnosis of HLRCC ;
2、Suggesting the efficacy of targeted therapy in patients with FH-deficient RCC.
| Detected Gene | Detected Region | Variant Type | Amplicon |
| FH | CDS | SNV、indel、CNV | 22 |
| MED12 | Exon1、Exon2、L1224F | SNV、indel | 4 |
PROJECT PARAMETER
| Project | Core Technology | Specification | Applicable Instruments | Sample Type |
FH and MED12 Gene Mutations Detection Kit | RingCap® | 16tests/kit 32tests/kit | Illumina MGIseq | Peripheral Blood, tissue (Pairing detection recommended) |
TARGET POPULATIONS
1. Uterine leiomyoma patients who initially test positive for FH-related immunohistochemistry.
2. Uterine leiomyoma patients who are young and have severe symptoms.
3. Young patients with type II pRCC.
4. Patients with multiple cutaneous leiomyomas and accompanying pain.
5. Patients with a family history of Hereditary Leiomyomatosis and Renal Cell Cancer (HLRCC) or close relatives of such individuals.
FEATURES & ADVANTAGES
1. Specialized Focus: Designed for common gene mutations associated with uterine leiomyomas, assisting in the diagnosis of HLRCC, and making it widely accessible.
2. Comprehensive Testing: Covers the entire coding sequence (CDS) of the FH gene, simultaneously detecting mutations and copy number variations (CNV), and also examines the most prevalent MED12 gene mutations in uterine leiomyomas.
3. Simple Operation: Based on the independent patent technology RingCap®, requiring only two steps for library construction.
4. Strict Quality Control: Strict quality control standards are implemented throughout the entire process.
DETECTION PROCESS
1. Nucleic Acid Extraction
2. Library Preparation (3.5 hours total time)
3. Sequencing
4. Auto-data Analysis
5. Report
[1] BJOG. 2017 Sep;124(10):1501-1512.
[2] N Engl J Med. 2013 Jul 4;369(1):43-53.
[3] WHO Classification of Tumors of the Female Reproductive System, 5th Edition
[4] Fertil Steril. 2014 Oct;102(4):1137-42.
[5] Nat Rev Dis Primers. 2016 Jun 23;2:16043.
[6] Chinese expert consensus on the diagnosis and treatment of fumarate hydratase-deficient uterine leiomyoma (2023 edition)
[7] Consensus on the clinical diagnosis and treatment of fumarate hydratase-deficient renal cell carcinoma
[8] NCCN Clinical Practice Guidelines in Kidney Cancer 2024.v1