Current location: Home > NEWS > Industry news
NEWS
PRODUCTS
Why is it not recommended to only detect POLE hotspot mutations, MMR and p53 immunohistochemistry?
News source: Release time:[2021-12-16]
Foreword
A few days ago, the Gynecological Oncology Committeeof the Chinese Anti-Cancer Association, the Pathology Branch of the ChineseMedical Association, and the International Pathology Quality Control Centerjointly issued the "Chinese Expert Consensus on Molecular Detection ofEndometrial Cancer (2021 Edition)" in the "Chinese Journal ofCancer". , Aims to improve the understanding of Chinese clinical workerson molecular detection of endometrial cancer, so as to guide and standardizethe clinical application of molecular detection of endometrial cancer in China.The consensus compares the differences of multiple detection methods andprovides corresponding reference materials.

Previously, many people in the industry suggested tous, why does Spacegen’s Human Endometrial Cancer Molecular Classification GeneMutation Detection Kit need to test so many genes? According to the NCCNguidelines, the detection of POLE hotspot mutations, MMR and p53immunohistochemistry can be enough for classification, and the cost would below. Here we discuss the following together with the newly released expertconsensus.
#01
Hot spot mutations cover mostbut not all POLE pathogenic sites
Expert consensus recommendations for POLE mutationdetection include hot spot mutation detection (category 2A) or POLE geneexonuclease domain (EDM) pathogenic mutation detection (category 2B). It istrue that the five common hotspot mutations P286R, V411L, S297F, A456P andS459F have covered 95.3% of the known pathogenic variants of POLE genes. Theconsensus still recommends the use of high-throughput sequencing methods todetect pathogenic mutations in exons 9-14 of the POLE gene.
The consensus refers to a study that systematicallyevaluated the pathogenicity of POLE mutations. Researchers analyzed the data of82 endometrial cancers with POLE somatic mutations in the TCGA study, and foundthat POLE EDM pathogenic mutations have the following specific genomic changes:(1) C>A conversion >20%, ( 2) T>G conversion>4%, (3) C>Gconversion <0.6%, (4) indels <5%, (5) TMB>100 mut/Mb.The researchers developed aPOLE pathogenicity scoring system. Each of the above genomic features is 1point, and 1 point is added for repeated occurrences in the TCGA or COSMIC ECdatabase.The researchers used a score of 4 as the cut-off value. In addition to 5hotspot mutations, tumors with a score of 4 or higher also carry 6 othernon-hotspot POLE EDM mutations. All of these mutations are recurring mutationsin the TCGA or COSMIC EC database. . Another 4 mutations with a score of 3 wereclassified as mutations of uncertain significance.
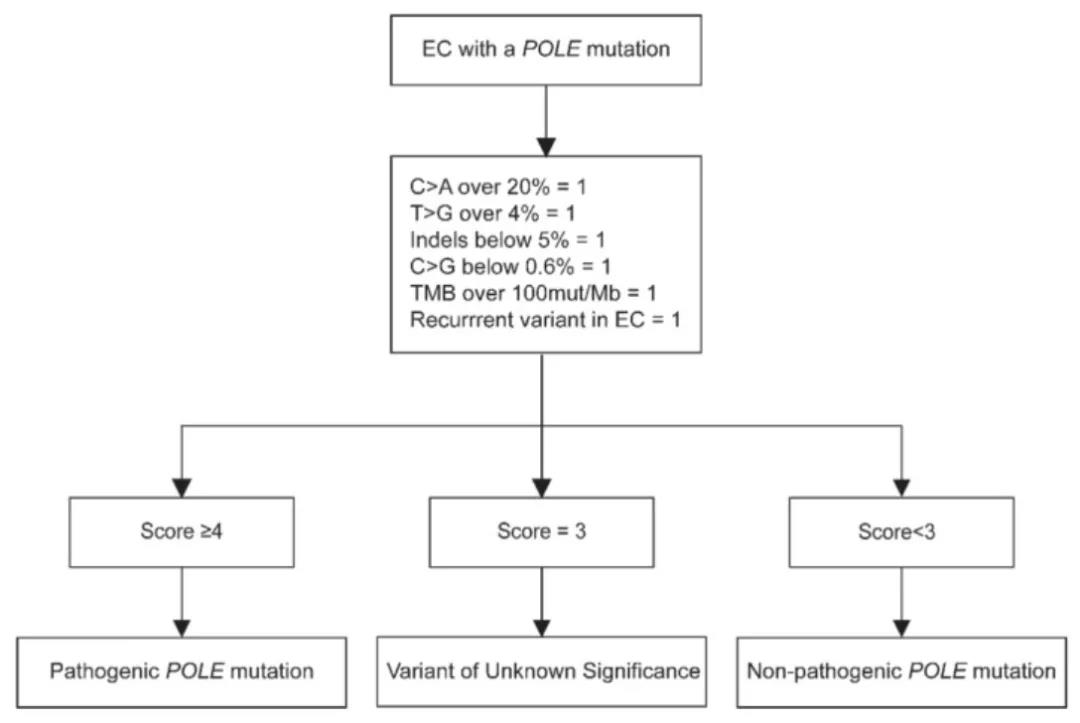
POLE Pathogenicity Score
In a cohort analysis of 3361 endometrial cancerpatients, the researchers found that patients with possible disease-causingmutations judged by the scoring system had a 5-year RFS of 92.3%, which wassimilar to the previously reported prognosis of the POLE hypermutation subtype,indicating that this part of patients should be classified as POLE hypermutant.
Although the scoring system is currently for referenceonly, it is undeniable that as the research continues to be deepen, thepathogenicity classification of many mutations will change. If only hotmutations are detected now, important information is missed, and it cannot betraced back and corrected in the future.
#02
Routine MMR/MSI screening maymiss Lynch syndrome patients
The original consensus mentioned that “a research teamused tumor single-tissue sequencing to screen 465 patients with stage IVcolorectal cancer for Lynch syndrome, with a sensitivity of 100% and aspecificity of 95.3%, while conventional MMR/MSI testing combined BRAF V600Escreening misses 10% of Lynch syndrome patients."
The study is part of the Ohio Colorectal CancerPrevention Program. A total of 465 patients were enrolled, including 46 knownLynch syndrome cases in the validation cohort and 419 members in the prospectivecohort. There were 12 cases of Lynch syndrome in the prospective cohort. UsingMSI or IHC for testing, followed by BRAF V600E testing, missed 5 and 6 patientswith Lynch syndrome. Compared with the sensitivity of 89.7% (95% CI78.8%-96.1%, P=0.04) for IHC plus BRAF and 91.4% (95% CI 81.0%-97.1%, P=0.07)for MSI plus BRAF, Tumor single sample sequencing has better sensitivity (100%,95% CI 93.8%-100%). The specificities of the three methods are relativelyclose.
The study found that in a patient with a prospectivePMS2 germline mutation, the MLH1 promoter had been methylated. According to theexisting screening methods, the case will be regarded as a sporadic tumor,genetic counseling and follow-up genetic testing are not recommended.
The article also mentioned that, according to thestandard screening process, the detection rate of MMR gene germline mutationsin dMMR patients without MLH1 promoter methylation is only 25% to 67%. Whengermline mutations are not detected, patients can only be diagnosed withunexplained MMR defects. However, up to 68% of unmethylated dMMR cases withoutgermline mutations are due to the 2 somatic mutations in the MMR gene. Usingtumor single-sample sequencing can better distinguish Lynch syndrome fromnon-Linch syndrome patients. In addition, the study also found that 380 samples(81.7%) had at least one somatic mutation that may be of therapeuticsignificance.
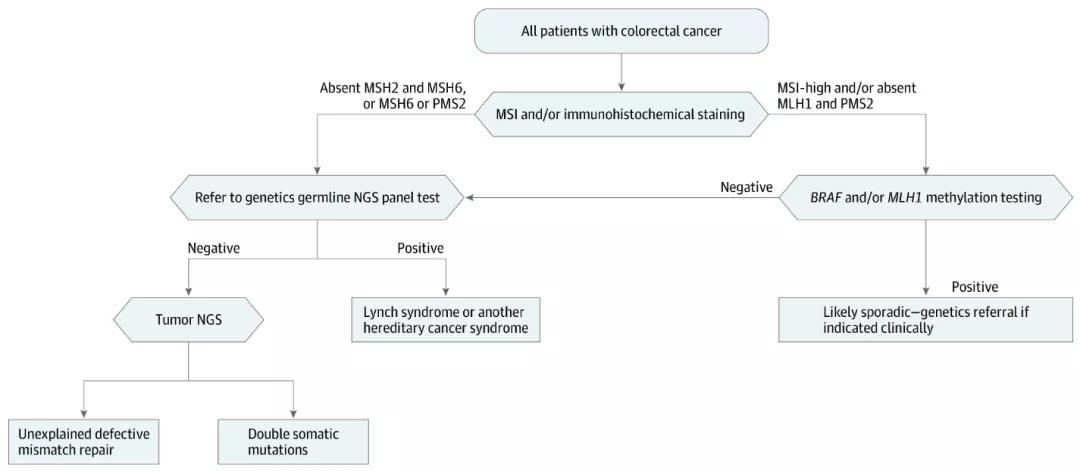
NGS According to the existingscreening model, unexplained MMR defects still require additional testing oftumor samples NGS
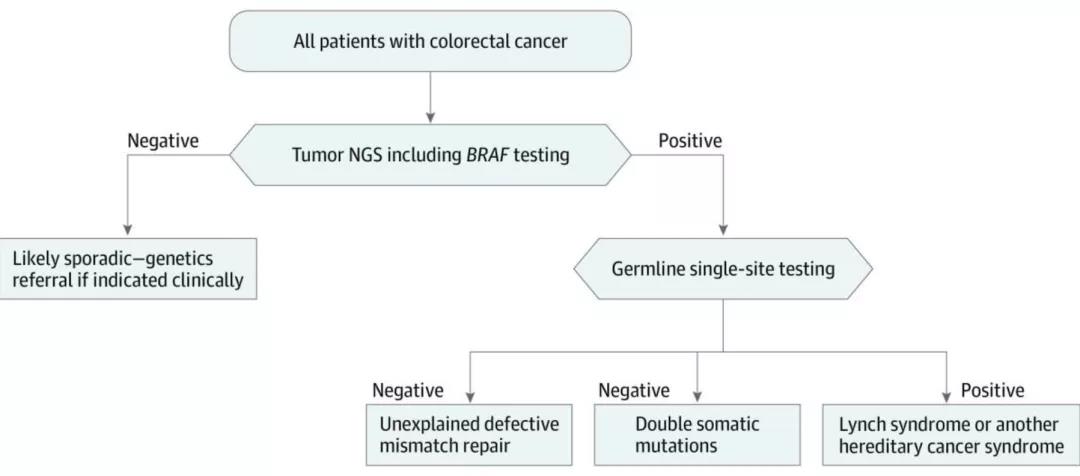
Research suggests using NGSfor initial screening of Lynch syndrome
Although this is a study based on patients withcolorectal cancer, considering the correlation between Lynch syndrome andendometrial cancer, this expert consensus also refers to the results of thestudy. It is also suggested that “when screening for Lynch syndrome in newlydiagnosed endometrial cancer patients, high-throughput sequencing methods canbe considered instead of conventional MMR/MSI detection, to detect multi-genesin high depth (without paired normal samples)." The genes tested includeMLH1, MSH2, MSH6, PMS2, EPCAMI.It is recommended to supplement POLE and TP53gene detection, which can be classified at the same time as the screening.
#03
There may be errors in P53 immunohistochemicalclassification
The original article of the expert consensus pointedout that "In the TCGA study, TP53 gene mutations were common in the copy-numberhigh group, and the incidence was about 92%. Therefore, TP53 gene mutation canbe used to replace copy-number high classfication, the consistency of p53protein immunohistochemical detection and TP53 gene mutation can reach 92.1%.It is easier to use p53 protein immunohistochemical detection in clinicalpractice, but it should be noted that this may result in about 15% of thecopy-number high population being classified into the NSMP group. "
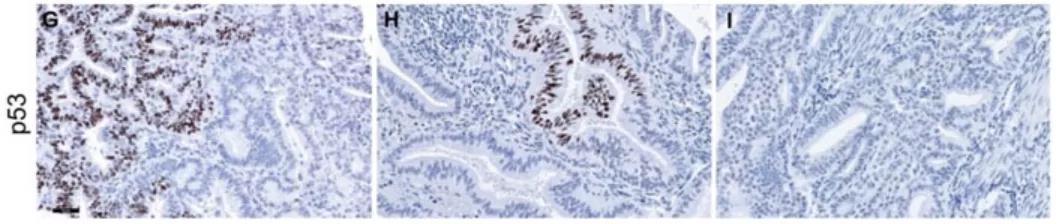
A reanalysis of TCGA data showed that up to 22% ofendometrial cancers have frameshift mutations or nonsense mutations in the TP53gene. These mutations will not result in the stable expression of p53 proteinand cannot be detected as p53 overexpression by IHC. Frameshift or nonsensemutations are associated with loss of p53 expression and may be misidentifiedas wild-type p53.
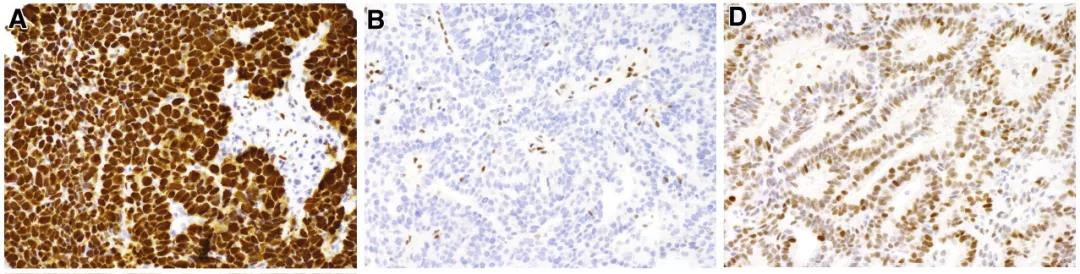
A.p53 overexpression, B. p53 expression iscompletely absent D.P53 wild type
(The actual case may not be astypical as the picture above)
In addition, the results ofimmunohistochemistry depend on staining and interpretation, some studies andliterature reported that p53 IHC and TP53 sequencing are inconsistent.
Inconsistent p53 expressionhas the same pathogenic TP53 mutation (c. 637C>CT)
#04
More and more immunological and targeted therapybiomarkers
Immunotherapy has been approved for the treatment ofpatients with recurrent or metastatic endometrial cancer. Currently approveddrugs are mainly monoclonal antibodies against PD-1 targets, such aspembrolizumab, nivolumab and dostarlimab-gxly. The approved molecular markersfor companion diagnostics are dMMR, MSI-H and TMB-H.
The FDA-approved drug targets for endometrial cancerinclude NTRK and HER2, some pan-cancer clinical studies have explored thetargeted therapeutic effects of genes such as PI3K/AKT/mTOR, KRAS, AKT1, FGFR2,FBXW7, and PTEN.
Consensus recommends that patients with recurrent ormetastatic endometrial cancer may consider using high-throughput sequencing todetect the fusion of TMB, MSI, and NTRK. The test samples are tumor tissue andmatched normal specimens (peripheral blood). It can detect PIK3CA, KRAS, AKT1,FBXW7, PTEN and other gene mutations, FGFR2 rearrangement or fusion, and HER2amplification at the same time, seeking cross-cancer drug indications andpan-cancer clinical trials.,as well as performing molecular classificaion.
Conclusion
In recent years, the development of high-throughputsequencing technology has deepened our understanding of the pathogenesis andmolecular genetic for endometrial cancer. The personalized and precisetreatment based on molecular genetic has revolutionized the treatment model ofendometrial cancer." With the deepening of research, the development oftechnology, and the reduction of sequencing costs, a more panoramic endometrialcancer detection panel will be the general trended.
Human Endometrial Cancer Molecular Classification GeneMutation Detection KitSpacegen's molecularclassification test for endometrial cancer is jointly developed with theDepartment of Pathology, Obstetrics and Gynecology Hospital of FudanUniversity. In addition to the mandatory POLE 9-14 exon, MSI and TP53 full CDSarea, it also covers MMR And EPCAM gene full CDS region, as well as CTNNB1,PIK3CA, KRAS, PTEN and other expandable genes.
Reference
[1] Chinese Expert Consensuson Molecular Detection of Endometrial Cancer (2021 Edition)
[2] J Pathol. 2020Mar;250(3):323-335.
[3] JAMA Oncol. 2018 Jun1;4(6):806-813.
[4] J Pathol. 2020Mar;250(3):336-345.
[5] Oncotarget. 2017 Apr11;8(15):25542-25551.
[6] Int J GynecolPathol. 2016 Jul;35(4):289-300.