Current location: Home > NEWS > Industry news
NEWS
PRODUCTS
Quantitative detection of free circulating tumor HPV in plasma
News source: Release time:[2024-09-30]
Foreword
Cervical cancer is a malignant tumor that seriously endangers women's health. According to the statistics of the National Cancer Center, there were 150,000 new cases of cervical cancer and 55,000 deaths in China in 2022, and the number of cases and deaths exceeded the sum of other gynecological tumors [1]. Early cervical cancer (≤ IB2 Stage) is usually treated by surgery, and the risk of recurrence is 8%-10% within two years; Radiotherapy and chemotherapy are adopted for the advanced stage, and the risk of recurrence within two years is 30%-50%. Routine clinical examinations are routinely performed in women who have been treated for invasive cervical cancer, with ancillary imaging studies such as magnetic resonance imaging (MRI) or computed tomography (CT) scans, if necessary. However, there are no reliable monitoring markers to predict which patients are more likely to relapse after cervical cancer treatment. Plasma free tumor HPV(HPV ctDNA or ctHPV) is a potential biomarker [2].
ctHPV and MRD(Molecular Residual Disease)detection
HPV is an epitheliophilic, non-enveloped, double-stranded circular small DNA virus with an approximate 8 kb genome that is divided into three regions: the early genes (containing open reading frames E6, E7, E1, E8, E2, E4, E5, 4.5kb), the late genes (containing open reading frames L2 and L1, 2.5kb), and the non-coding upstream regulatory region (URR), also known as the long control region (LCR, 1kb). HPV genomic sequences are more than 10% different, i.e. different genotypes. At present, more than 200 HPV types have been found, isolated and identified, and divided into high-risk and low-risk types according to their cancer-inducing potential. HPV type 16/18 accounts for about 70%[3].
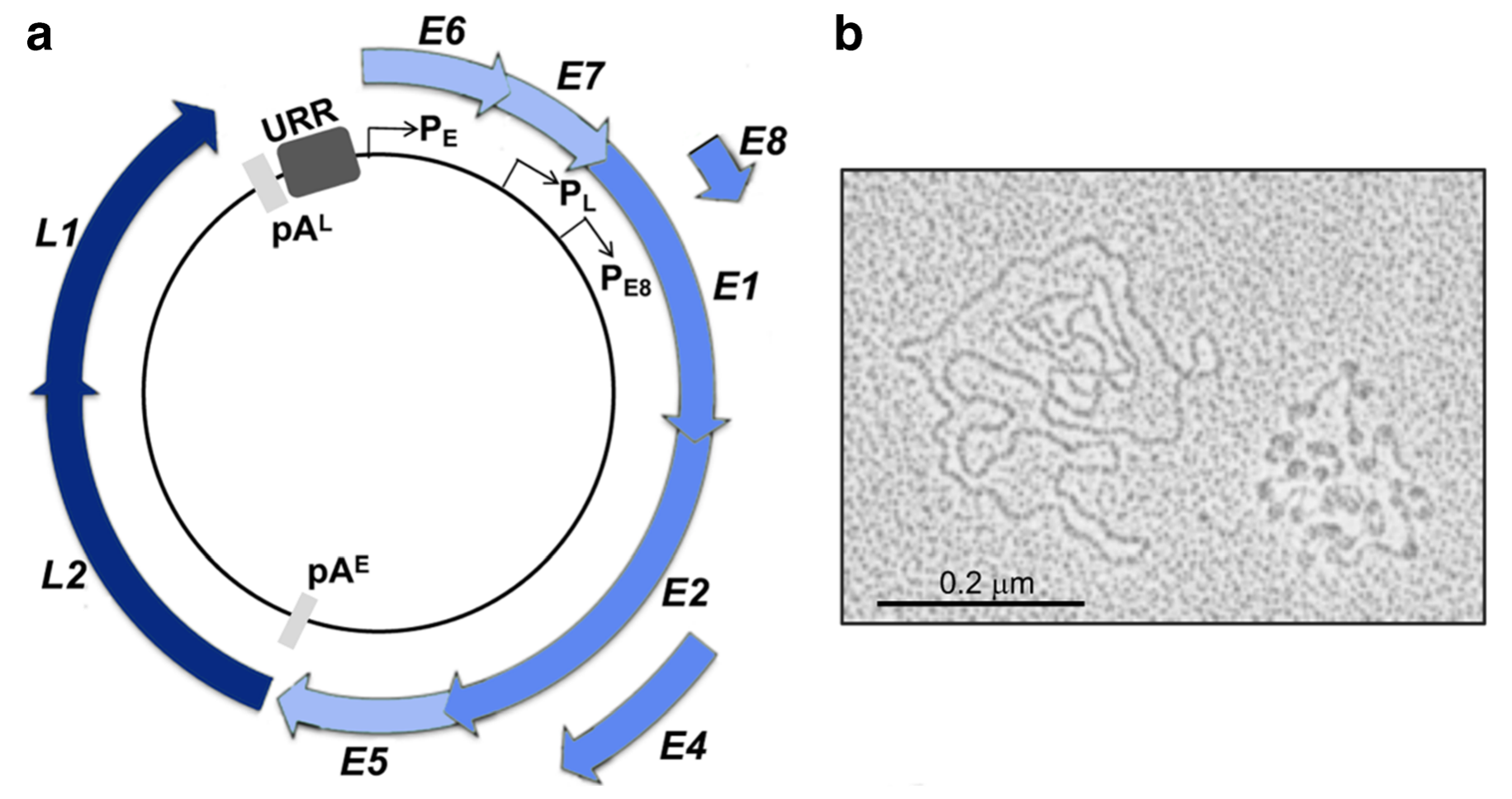 ▲HPV virus genomic structure and HPV molecules under electron microscope [4]
▲HPV virus genomic structure and HPV molecules under electron microscope [4]
E6 and E7 are the most closely related oncogenes to cervical cancer. During the continuous infection of high-risk HPV(hrHPV), HPV DNA is usually integrated into the host genome and the negative regulatory factors E1 and/or E2 are destroyed, resulting in the disordered expression of viral oncogenes E6 and E7 [5]. The binding of E6 and E7 proteins to p53 and pRb, respectively, leads to a significant reduction in the level of cancer suppressor proteins in cells, unstable host chromosomes, and disordered DNA replication, thereby inducing tumors [2]. In other studies, Nanopore was used to analyze the integration of HPV16 and found that some viruses (E6/E7) were not integrated into the genome, but their proportion was very low [5].
Molecular residual lesions (MRD) refer to molecular abnormalities of tumor origin found in liquid biopsies such as blood by cell molecular biology methods, although traditional imaging examinations or laboratory tests cannot find MRD after treatment, suggesting that tumor cells are persistent or clinical progress is possible. CtDNA is a tumor genomic fragment released into the circulatory system mainly derived from primary tumors, metastases or circulating tumor cells, which carries tumor-specific genetic or epigenetic variation and can reflect the tumor state in vivo in real time. It is one of the most widely used biomarkers for liquid biopsy in clinical practice [6].
The allele frequency (VAF) of somatic mutations in ctDNA is very low, even in advanced/metastatic cancer (stage III/IV), the average VAF is 3.67%, and the median VAF is only 0.41%[7]. Therefore, higher sequencing depth is required to effectively improve the sensitivity of mutation detection [6]. Since HPV is the cause of most cervical cancer and exists in multiple copies in cells, it should also be released in the form of ctDNA, serving as a potential biomarker [2]. CtHPV is not disrupted by a high wild-type background in total cfDNA, and is easier to detect and identify than point mutations, without the need for customized procedures.
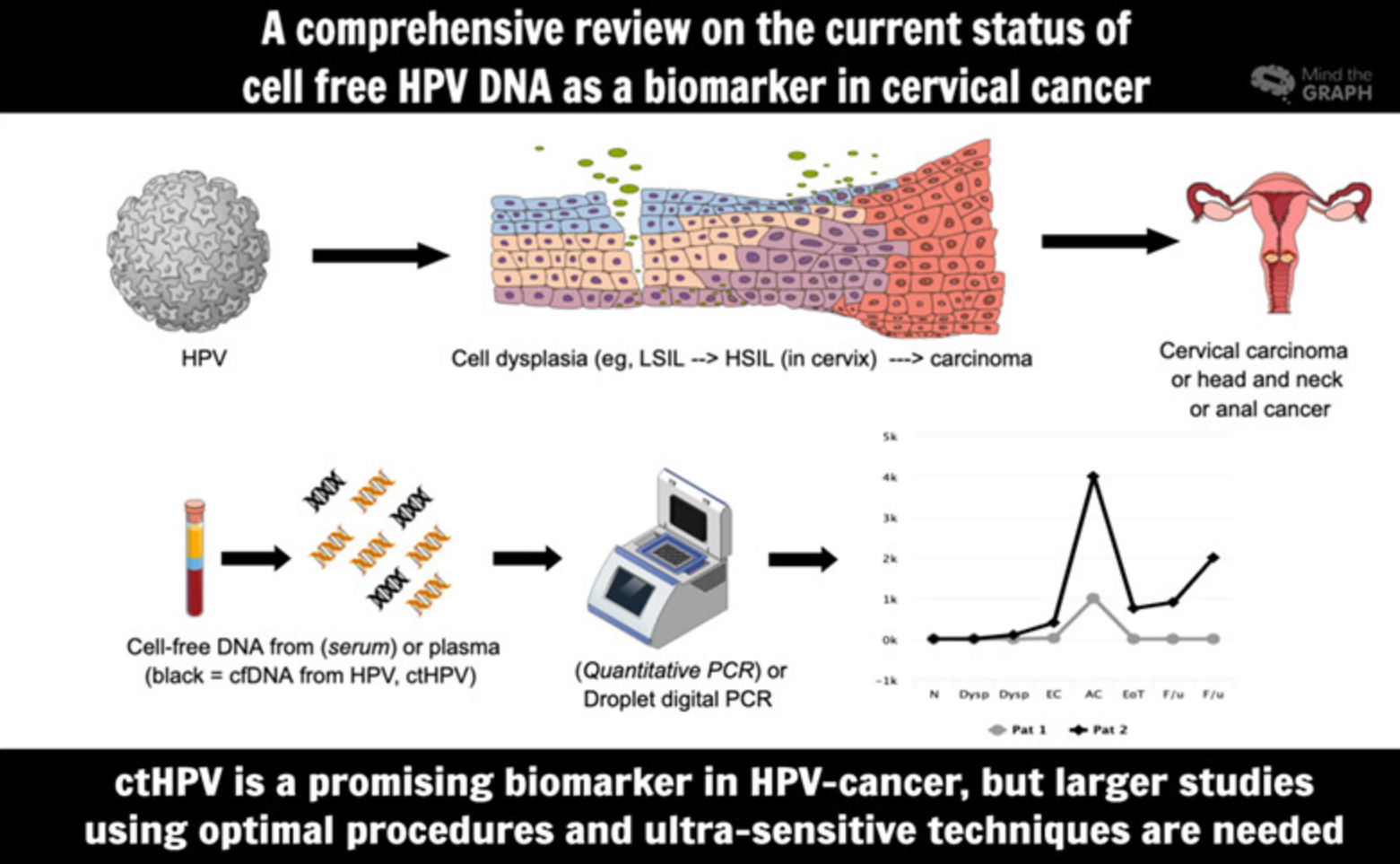 Bring about cervical cancer ctHPV detection overview [2]
Bring about cervical cancer ctHPV detection overview [2]
Application of ctHPV detection in cervical cancer
A prospective PAIR-HPV study (NCT02554565) enrolled 55 patients, predominantly in stage II-IVA, who received first-line treatment with radical chemoradiotherapy (CRT). HPV ctDNA was successfully detected in 69% of patients prior to CRT treatment. HPV ctDNA levels were correlated with the number of HPV copies in the tumor (r=0.41, p<0.001), tumor stage (p=0.037), and lymph node status (p=0.02). At the end of the CRT and during the follow-up period, HPV ctDNA positivity was associated with lower disease-free survival (DFS, p=0.048) and overall survival (OS, p=0.0013) [8].
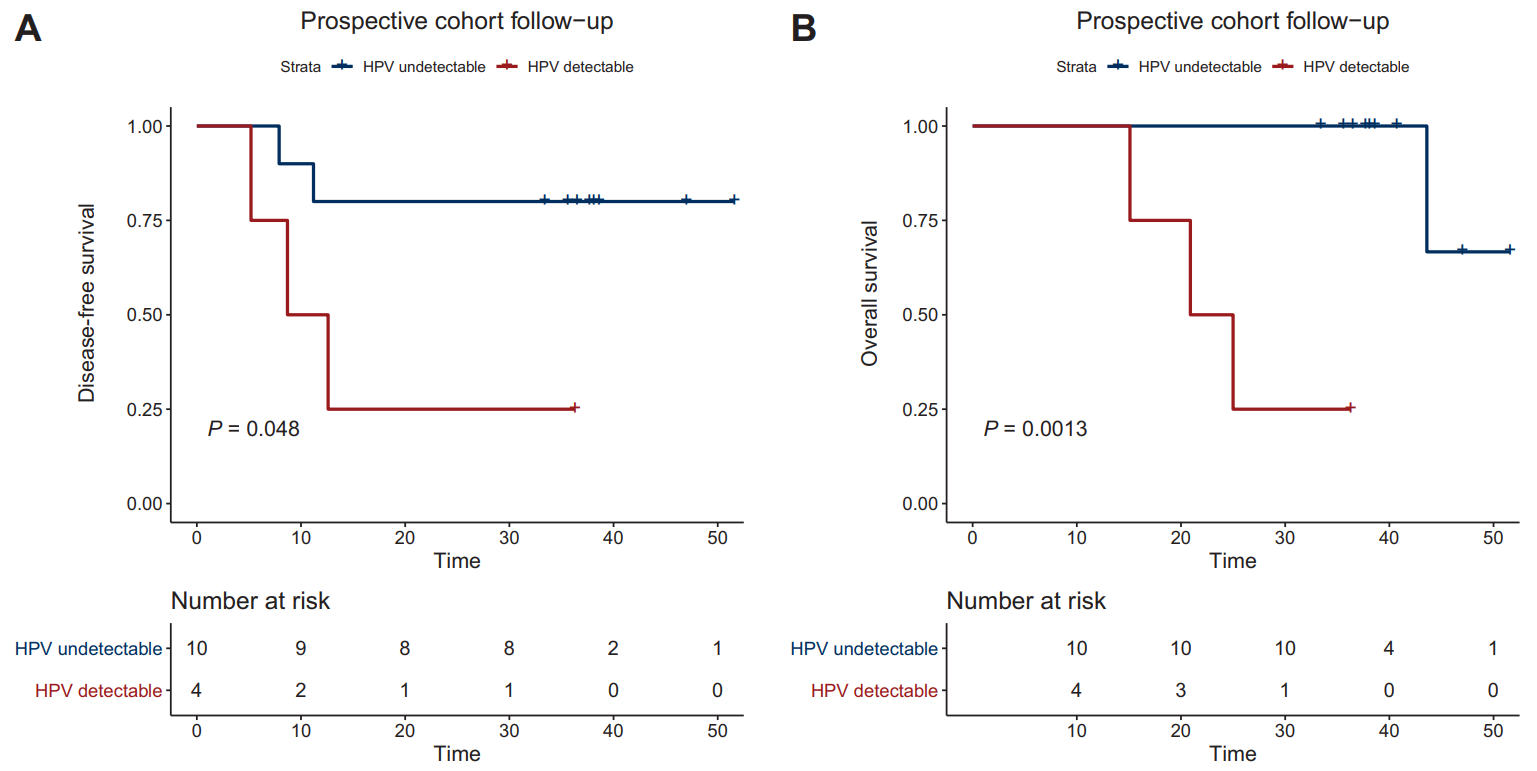 ▲ctHPV test results suggest that the prognosis of patients [8]
▲ctHPV test results suggest that the prognosis of patients [8]
A BioRAID study (NCT02428842) included 419 patients and 94 were analyzed for progression-free survival (PFS). FIGO III/IV in 73% of patients, all type 16/18, of which 20% were treated with surgery, 65% with chemoradiotherapy, and 15% with neoadjuvant therapy. The HPV E7 gene was used as a marker in the study. HPV ctDNA was detected in 63% of patients prior to treatment. HPV ctDNA positivity was associated with a high FIGO stage (p=0.02) and paraaortic lymph node involvement (p= 0.01). HPV ctDNA levels positively correlated with HPV copy numbers in the tumor (r=0.39, p<0.001)[9].
PFS was not associated with HPV ctDNA at baseline (HR=1.59;; p=0.19); Post-treatment HPV ctDNA positivity was associated with a shorter PFS (p<0.0001). End-of-treatment (EOT)/ follow-up process samples were collected from 44 follow-up patients for monitoring. With one exception, patients who were negative for EOT ctHPV remained negative during follow-up. Seventy-five percent of EOT ctHPV-positive patients remained positive during follow-up and all relapsed; Two patients who cleared ctHPV during follow-up did not relapse. The median lead time for detecting positive ctHPV is 10 months (2–15 months) compared with that for clinically diagnosed relapse [9].
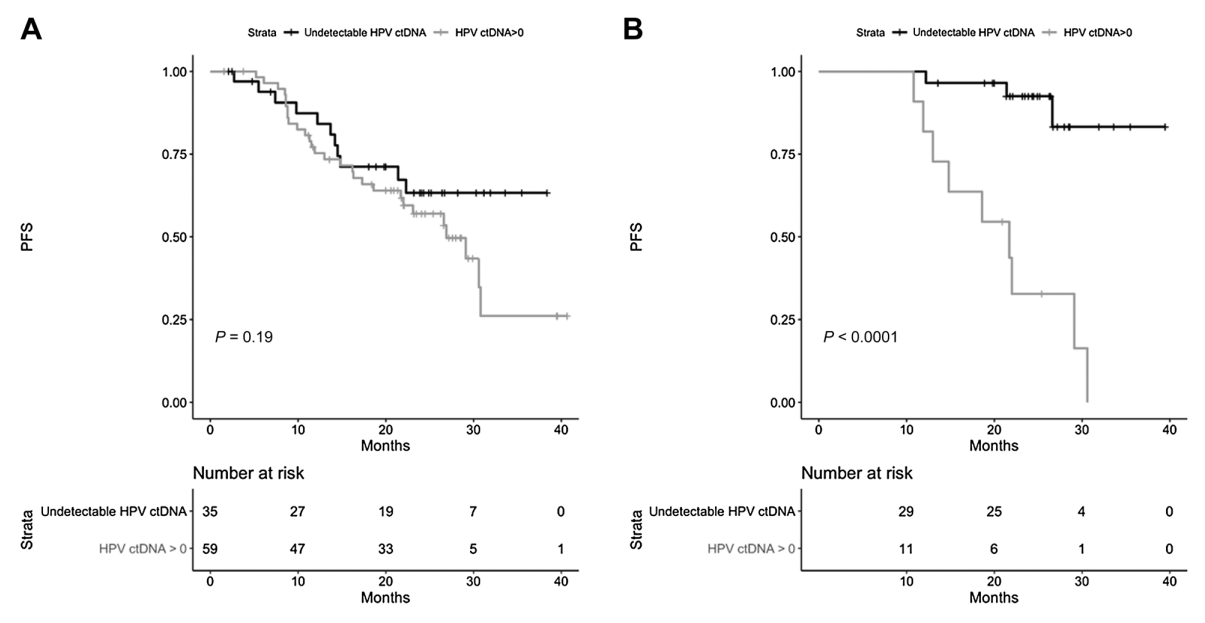 ▲ Relationship between ctHPV test results and PFS of patients (A, baseline test, B, EOT test) [9]
▲ Relationship between ctHPV test results and PFS of patients (A, baseline test, B, EOT test) [9]
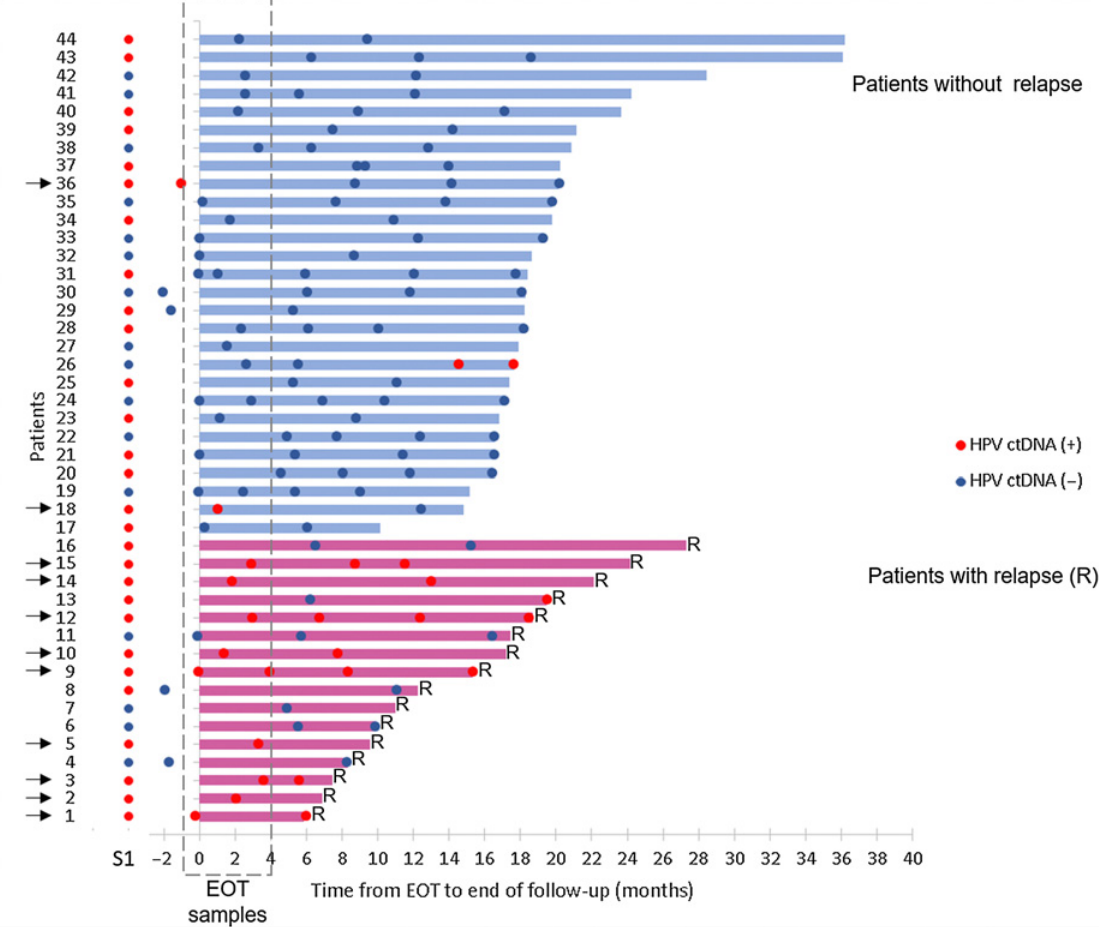 ▲ From EOT to the end of the follow-up, ctHPV monitoring results [9]
▲ From EOT to the end of the follow-up, ctHPV monitoring results [9]
A Swedish study involving 74 patients with locally advanced cervical cancer analyzed 418 plasma samples by digital PCR(ddPCR) with a median follow-up of 37 months. 92.4% of pretreatment samples were ctHPV positive. The presence of ctHPV in plasma was associated with poorer PFS (P < 0.001). At the early follow-up, the positive predictive value (PPV) for ctHPV status in predicting disease progression was 87.5% and the negative predictive value (NPV) was 89.3%. In all (10) patients who relapsed after a complete response, the median lead time for ctHPV detection was 315 days compared with imaging for recurrence [10].
Application of ctHPV detection in oropharyngeal cancer
In addition to cervical cancer, squamous cell carcinoma of the head and neck, anal cancer, vaginal cancer, vulvar cancer and other cancers are caused by HPV infection in a certain proportion. In recent years, the incidence rate of HPV infection in oropharyngeal cancer of Europe and America has increased significantly. Studies have shown that most of the cases have a direct relationship with HPV infection. China also has a trend of increasing year by year. Based on a meta-analysis of HPV16 testing, the overall infection rate of HPV16 in head and neck tumors in China is 24.7% and that in oropharyngeal cancer is 31.6%. A recent study based on WHO database showed that the proportion of HPV positive oropharyngeal cancer in China was 25.8%[11].
A Naveris-led study (NCT02281955, NCT03077243, NCT0316182) analyzed 509 blood samples from 45 patients with a median follow-up of 19.2 months. CtHPV was undetectable at all time points after treatment in 37 patients, and no patient relapsed, with an NPV of 100%. Eight patients had ctHPV positivity during posttreatment monitoring, four of which were confirmed by biopsy to have relapsed. These four patients had ctHPV positivity twice in a row, with a PPV of 100%. The median lead time between CT-HPV positivity and biopsy-proven recurrence was 6.6 months (3.3-12.9 months) [12].
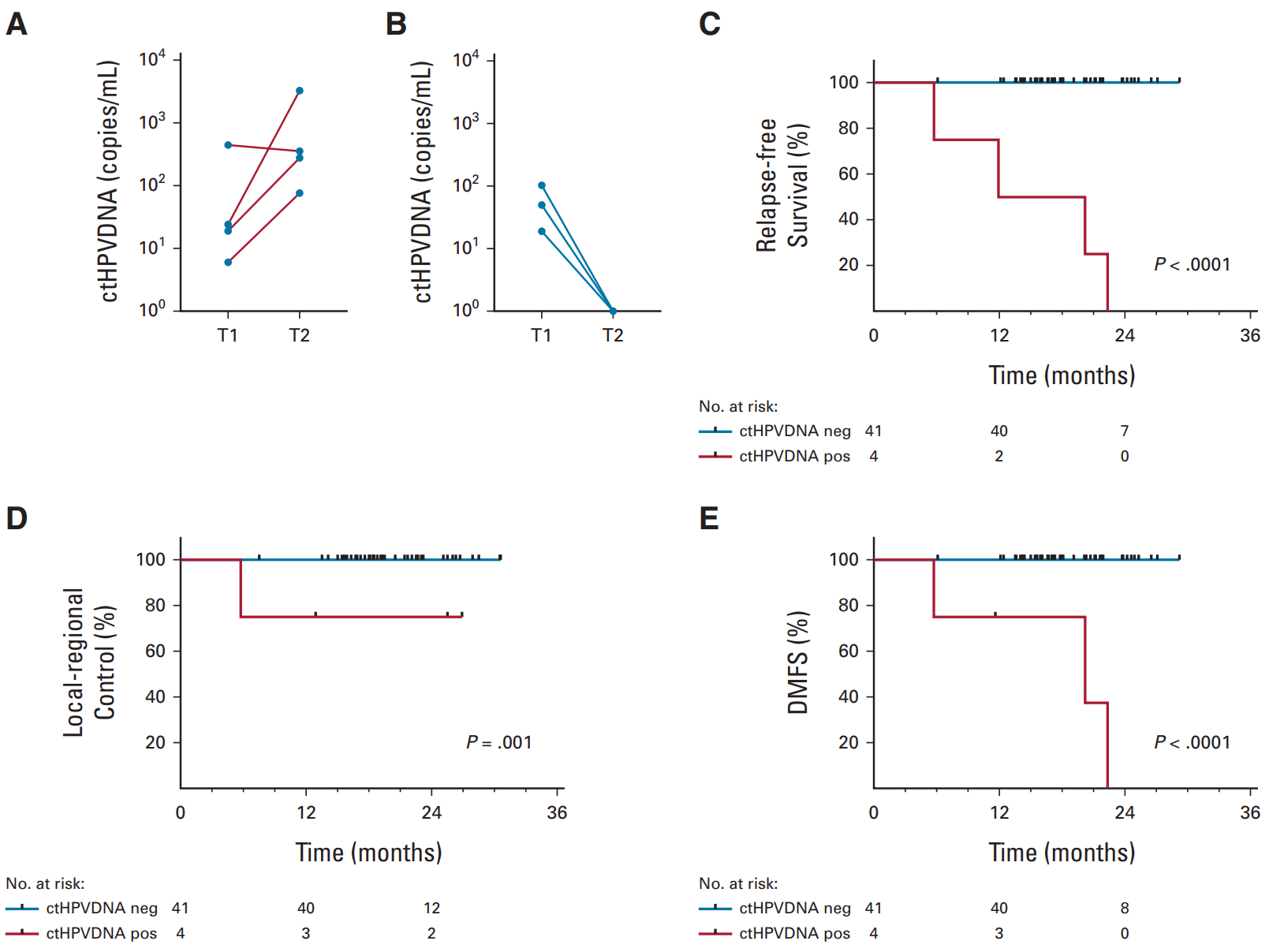
▲ Two consecutive ctHPV monitoring abnormalities in patients with higher risk of recurrence [12]
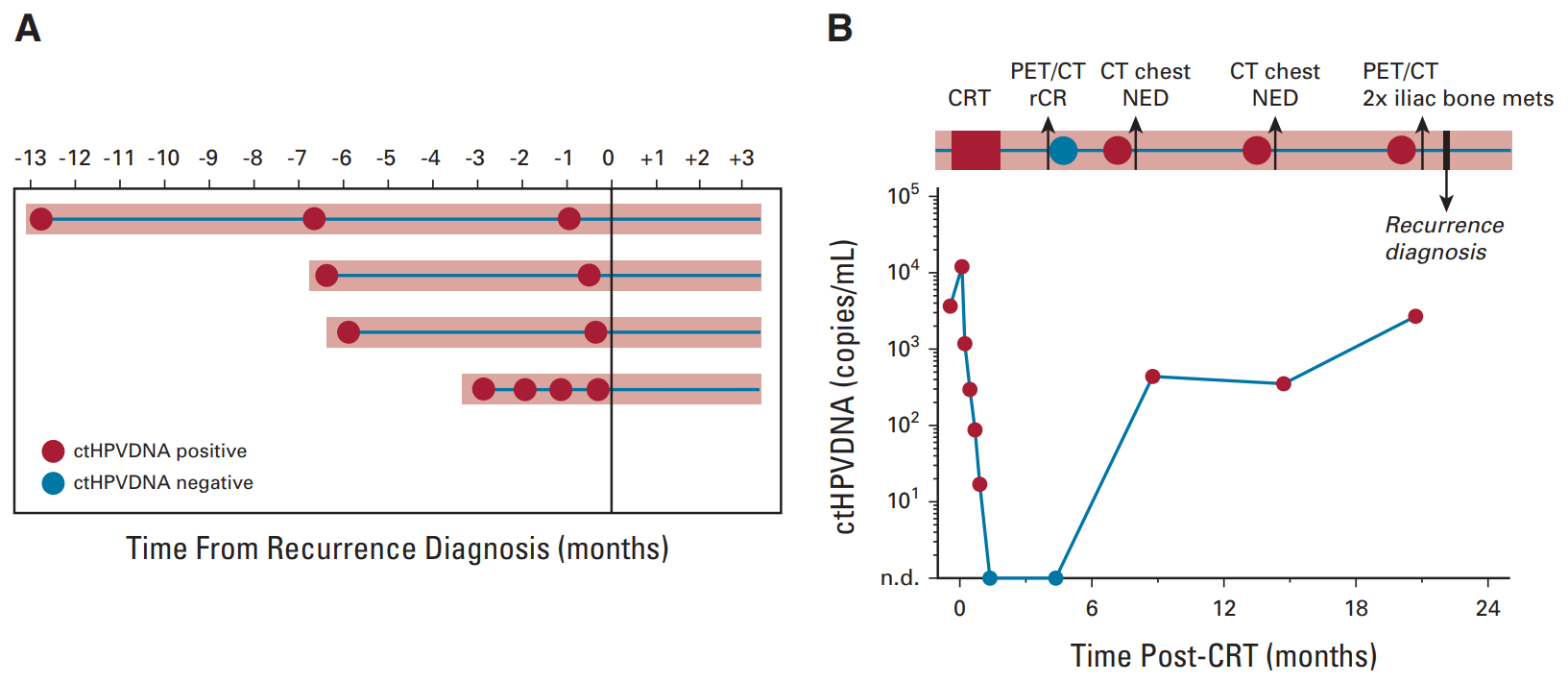 ▲ctHPV monitoring contributes to early detection of disease recurrence [12]
▲ctHPV monitoring contributes to early detection of disease recurrence [12]
A study in Japan quantitated ctHPV16 DNA by ddPCR in 35 patients with HPV 16-associated head and neck squamous cell carcinoma treated with radiotherapy with or without chemotherapy. After a median follow-up of 21 months, the NPV of ctHPV16 DNA and PET-CT were similar (89.7% vs 84.0%), while the PPV of ctHPV16 DNA was much higher than that of PET-CT(100% vs 50.0%)[13]. CSCO guidelines for the diagnosis and treatment of head and neck tumors also state in the notes that "For HPV-related oropharyngeal cancer, recent studies have shown that regular copy numbers (tests) of HPV DNA in peripheral blood after treatment contribute to early detection of tumor recurrence" [11]. It is believed that this monitoring programme will soon be included in the guidelines.
Detection methods and precautions of ctHPV
The early ctHPV studies have one thing in common, namely, the sensitivity is quite poor, and most of them are only 6.9%–30%. The reasons may be manifold. Fluorescent quantitative PCR(qPCR) is not sensitive enough to detect trace amounts of ctHPV. The amount of plasma samples collected, and the methods of storage and treatment also have an impact on the test results [2]. In 2016, Jeannot et al. published the first study using ddPCR to detect ctHPV DNA in plasma. In 47 cervical cancer patients with HPV16/18 positivity, they found ctHPV DNA in pretreatment plasma using ddPCR(16/18 E7) in 83% of cases, compared with 60% (p = 0.02) using qPCR [14].
The high-throughput sequencing (HPV-seq) targeting hpv is also one of the options. HPV-seq (12 high-frequency mutated genes in targeted HPV and squamous cell carcinoma) was found to reproduce a detection limit of less than 0.6 copy level in cell line data. HPV-seq and ddPCR results were highly consistent (r2=0.95, p=1.9×10-29). HPV-seq can detect ctDNA in ddPCR-negative plasma as low as 0.03 copies /mL post-treatment. HPV ctDNA positivity at the end of treatment was associated with poor PFS with a sensitivity of 100% and a specificity of 67% (both 75% by DD PCR) for detecting relapse [15].
A prospective, multicenter, validated study (NCT03853915, NCT03702309) enrolled 70 patients with stage IB-IVA cervical cancer receiving CRT. Peripheral blood was collected at baseline, at the end of the CRT, 4–6 weeks after CRT, and 3 months after CRT, and ctHPV levels were measured using both ddPCR and HPV-seq methods. The primary endpoint was the 2-year PFS rate, with a median follow-up of 2.2 years, and 24 PFS events in 70 HPV-positive cervical cancer patients. HPV-seq showed similar results to ddPCR. Compared to patients who were negative for ctHPV, patients who were positive for ctHPV had significantly worse PFS at CRT end, 4–6 weeks after CRT, and 3 months after CRT at 2 years. The median lead time to relapse compared to imaging was 5.9 months for both ddPCR and HPV-seq [16].
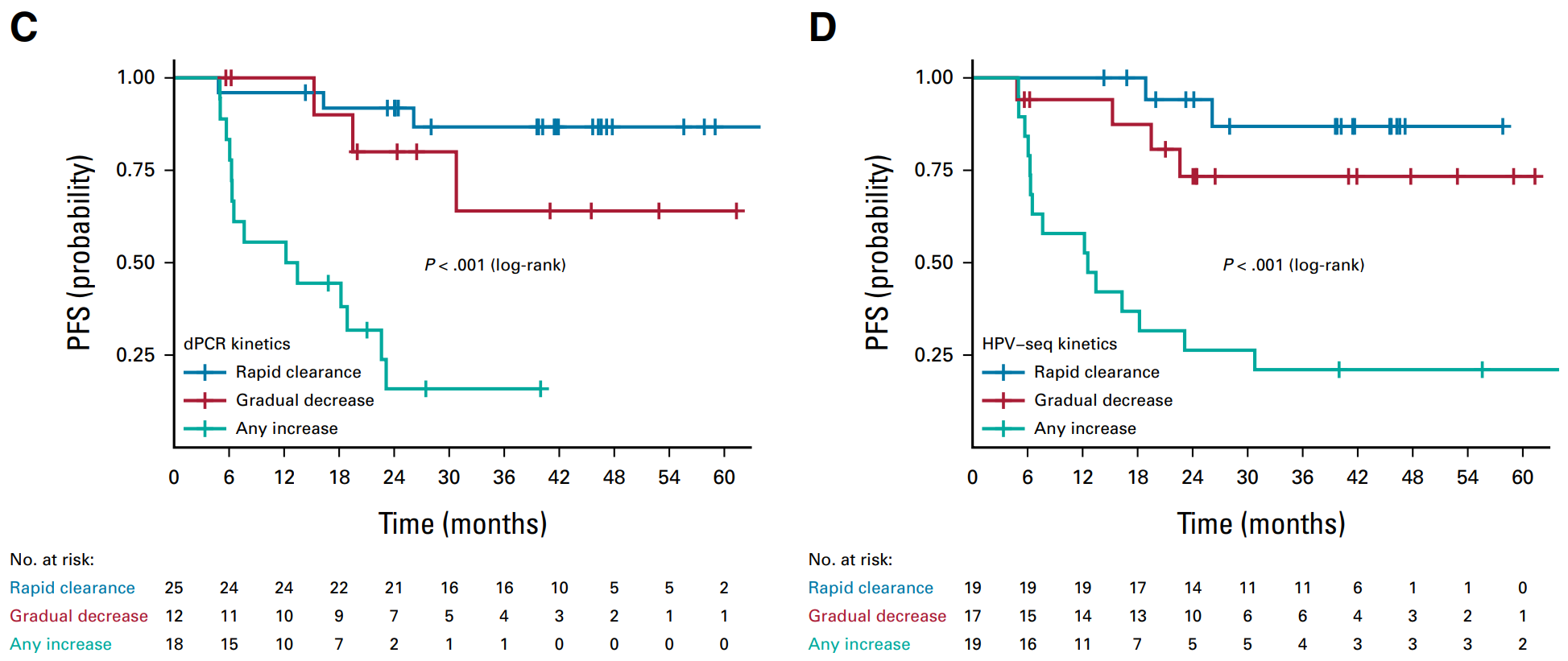
▲ These two test methods show consistent: patients with rapid removal of ctHPV have better PFS, followed by patients with gradual removal; PFS of patients with elevated ctHPV is significantly worse [16].
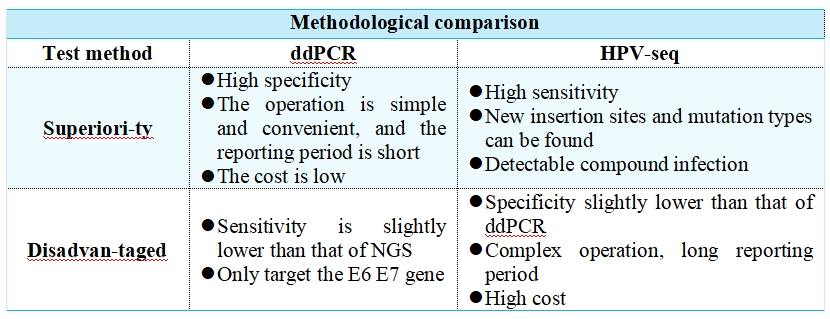
It should be noted that quantitative testing of ctHPV is not suitable for screening in healthy women or patients with precancerous lesions. CtHPV;was not detected in plasma of patients with precancerous lesions in nine studies using qPCR and in recent studies using ddPCR; In the only study detected, the association of HPV status in plasma with cervical samples was fairly poor (44%). The detection rate of ctHPV using qPCR or ddPCR in most healthy women is only 0% to 2%. The positive rate of ctHPV in early-stage patients is also low, with the baseline ctHPV positive rate of 24.1–65% in patients with stage I–II cervical cancer; The baseline ctHPV positive rate in advanced (stage III–IV) patients can reach 63–100% [2].
Summary
ctHPV tests the E6/E7 gene of HPV free DNA integrated into the human genome and released into plasma for molecular residual disease(MRD)detection, which can help predict therapeutic effects and even prompt disease recurrence several months in advance. The digital PCR method is adopted for quantitative detection of human ctHPV16/18 of SpaceGen, with the sensitivity up to 1–3 copies/reaction; besides, the experimental operation is simple, and the reporting period is short. Fuso Biological is committed to providing the most innovative products and services for the individualized and precise medical detection of tumors, and continuously updates the existing products.
References
[1] Zhonghua Zhong Liu Za Zhi. 2024 Mar 23; 46(3):221-231.
[2] Int J Cancer. 2023 Jun 1; 152(11):2232-2242.
[3] China Expert Consensus on the Use of Human Papillomavirus Nucleic Acid Detection for Cervical Cancer Screening (2022)
[4] Semin Immunopathol. 2020 Apr; 42(2):159-171.
[5] Nat Commun. 2022 May 10; 13(1):2563.
[6] China expert consensus on the detection and clinical application of molecular residual lesions in gastric cancer (2023 edition)
[7] Clin Cancer Res. 2018 Aug 1; 24(15):3528-3538.
[8] ESMO Open. 2021 Jun; 6(3):100154.
[9] Clin Cancer Res. 2021 Nov 1; 27(21):5869-5877.
[10] Clin Cancer Res. 2024 Jul 1; 30(13):2764-2771.
[11] CSCO guidelines for diagnosis and treatment of head and neck cancer 2023
[12] J Clin Oncol. 2020 Apr 1; 38(10):1050-1058.
[13] Int J Cancer. 2021 Feb 15; 148(4):995-1005.
[14] J Pathol Clin Res. 2016 Jun 28; 2(4):201-209.
[15] Clin Cancer Res. 2021 Nov 1; 27(21):5857-5868.
[16] J Clin Oncol. 2024 Feb 1; 42(4):431-440.
Statement:
This article is only for sharing, if it involves copyright issues, please contact us as soon as possible, we will correct the first time, thank you!