Current location: Home > NEWS > Industry news
NEWS
PRODUCTS
Fusion gene detection: how to choose DNA or RNA?
News source: Release time:[2024-09-30]
01. What is gene fusion
Gene fusion refers to the process in which some or all sequences of two genes are fused with each other to form a new gene. A percentage of the fusion genes are transcribed into chimeric fusion transcripts that can form fusion proteins by retaining the reading frame of their parent gene, affect parent gene expression, or act as long noncoding chimeric RNA.
In general, gene fusion refers to fusion at the genomic level. However, fusion may also occur at the transcriptome level, mainly because RNA produced by transcription of two different genes fuse together for some reason to form a new fused RNA, which may or may not encode a protein. The fusion genes generated at the genomic level may or may not be expressed (such as destroying the promoter region or other reasons) according to the fusion situation.
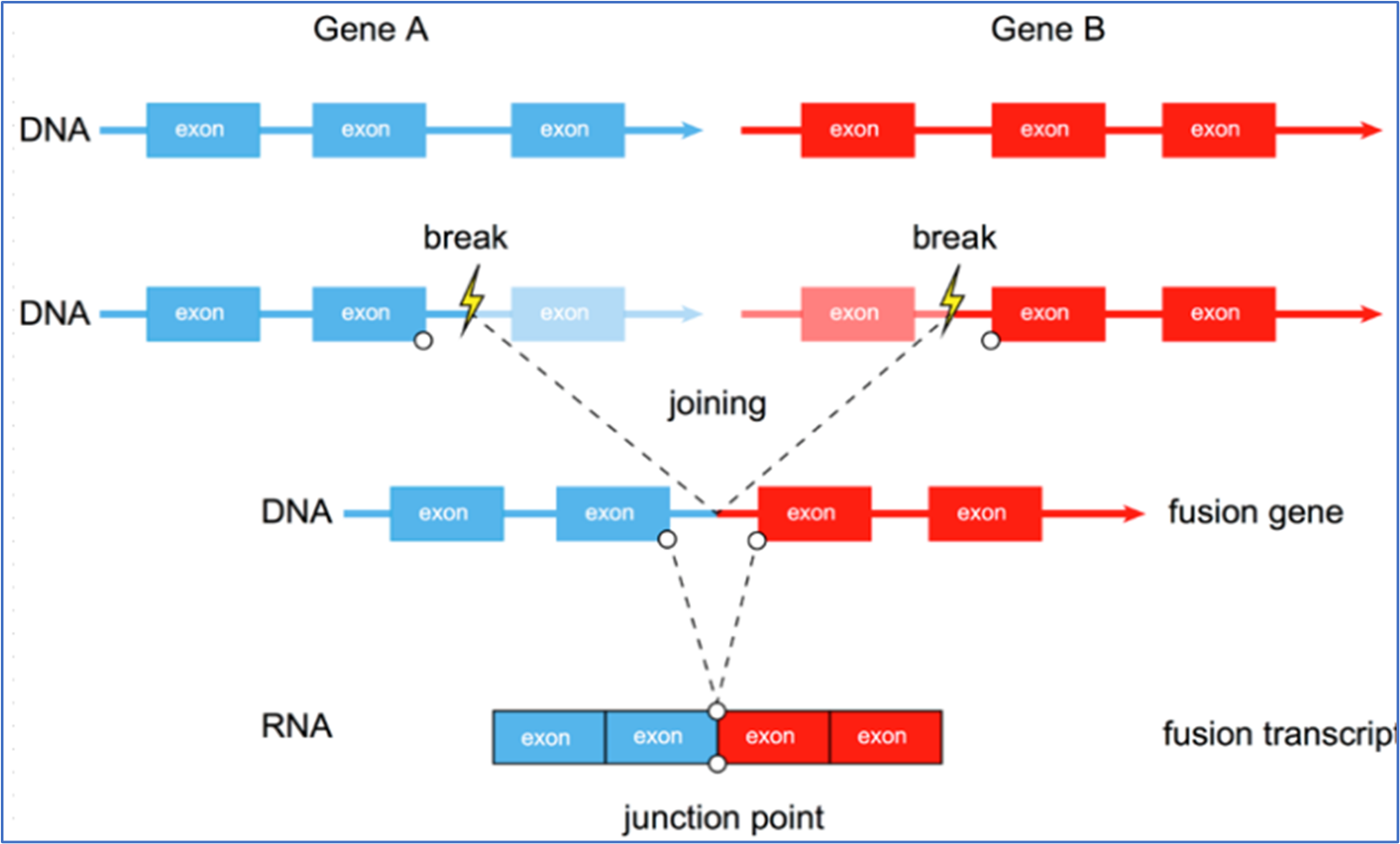 Schematic diagram of gene fusion
Schematic diagram of gene fusion
02. The production mechanism of fusion gene
Caused by rearrangement of chromosomal layers or by missplicing during transcription. The rearrangement at the DNA level mainly includes six forms:
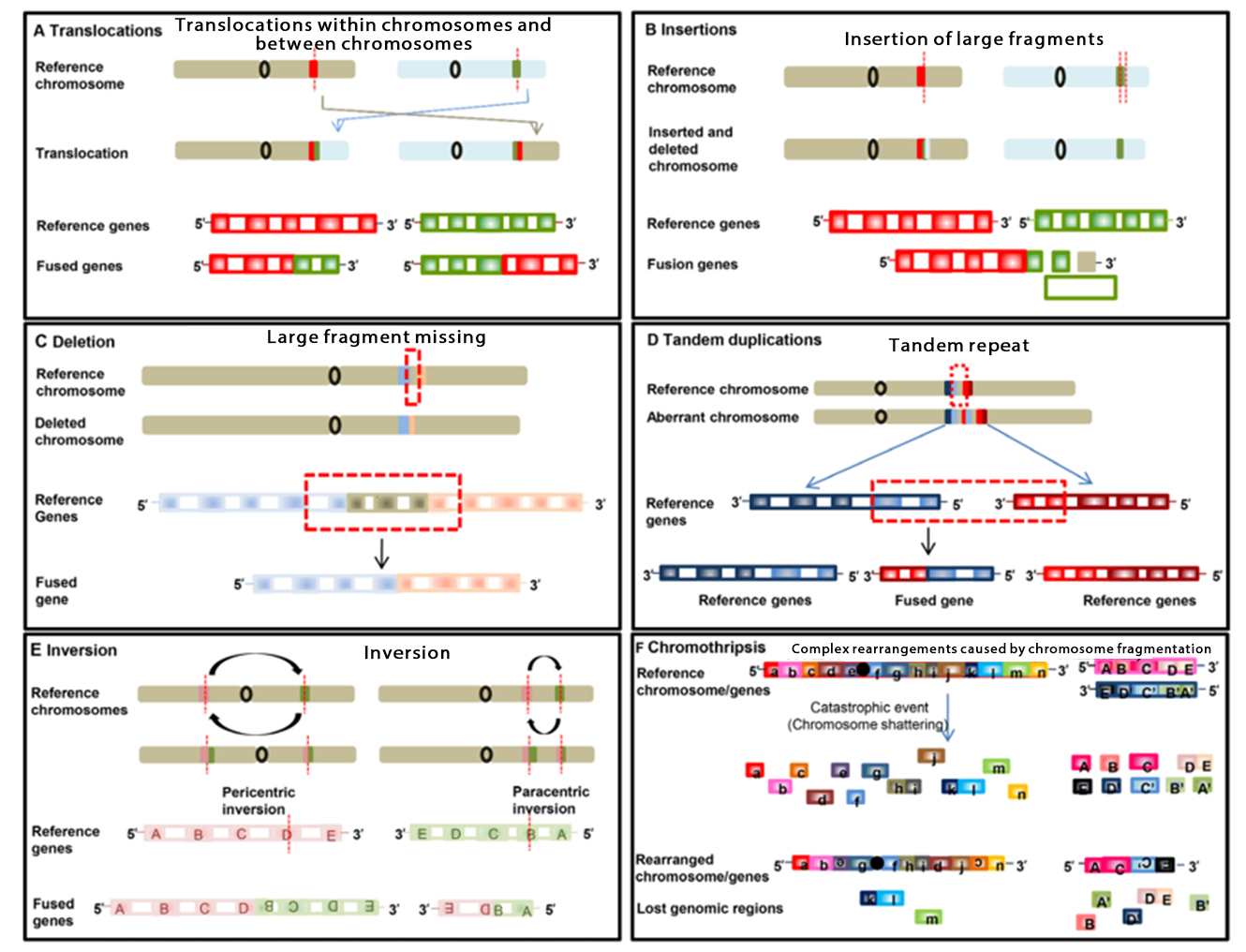 ▲ Rearrangement forms at DNA level
▲ Rearrangement forms at DNA level
At the RNA level, "read-through events" during transcription may also produce fusion proteins. When an RNA polymerase fails to properly terminate transcription at one gene end and continues transcription to the next due to abnormal splicing, a chimeric transcript is produced called a "read-through event" and a fusion transcript can be produced without involving rearrangement of genomic material (DNA rearrangement).
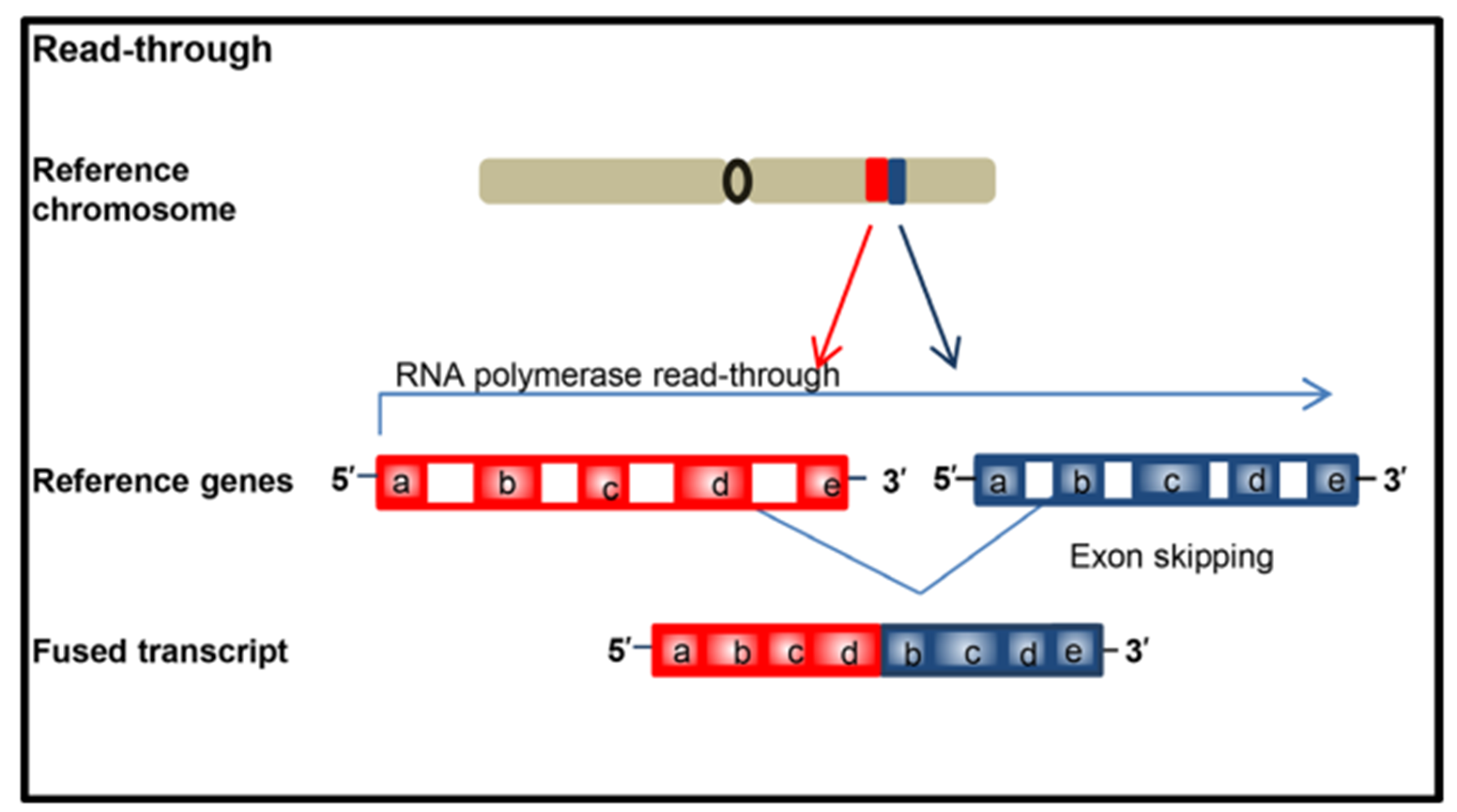 ▲ The "read-through events" in the transcription process
▲ The "read-through events" in the transcription process
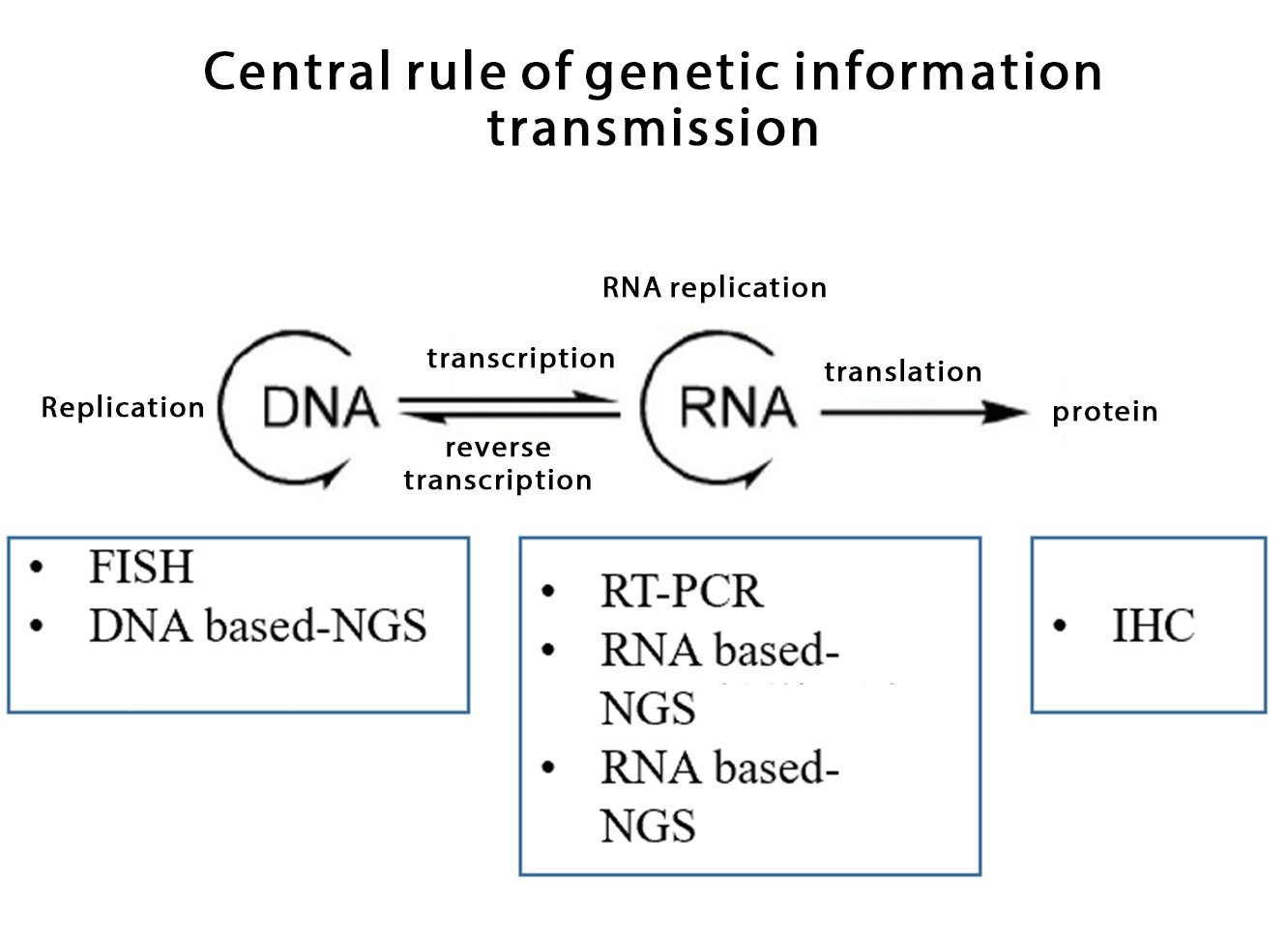
03. Common detection method of fusion gene
FISH
A molecular detection technique that hybridizes a fluorescence-labeled probe with a DNA target sequence in the nucleus, and observes and analyzes gene amplification or fusion variation under a fluorescence microscope is generally considered as a "gold standard" for detecting gene amplification and fusion, but it also has certain limitations. If not all detected fusions can produce expressible fusion RNA, the separation probe may miss a small intrachromosomal rearrangement resulting in a false negative result, and the different subtypes of fusion gene variation cannot be identified and distinguished, etc.
IHC
A technique for detecting the expression of a specific protein in a tissue or cytological section using antigen-antibody reactions and chemical coloration. The IHC detection has the advantages of high sensitivity, low cost, short detection time and easy automation. However, its accuracy depends on the presence of verified antibodies to the detected fusion proteins. For example, VentanaD5F3 IHC has high accuracy in detecting ALK fusion in lung adenocarcinoma patients, while ROS1 and NTRK positive IHC results need to be verified by other detection methods.
RT-PCR
The detection of fusion RNA at RNA level has high sensitivity and short detection time, but it is limited to the known fusions within the design range of detection primers, and the unknown fusions cannot be detected. In addition, the accuracy of the test results by this method is highly dependent on the quality of RNA in the sample.
Targeted second generation sequencing
By a large-scale parallel sequencing method, a plurality of fusion genes are simultaneously detected on the region within the design range of the primer or the probe. Targeted second-generation sequencing detects gene fusion at the DNA or RNA level, depending on the nucleic acid input.
According to the central principle, mature RNA is produced through transcription and splicing processing using DNA as a template. At the DNA level, the location of fusion breakpoints usually occurs in relatively long intron regions, and the fused breakpoints may be different for different patients, so it is difficult to directly amplify the breakpoints by traditional PCR. It is a feasible detection method to design probes to capture the breakpoints by using NGS method, but the detection of fusion genes at the DNA level inevitably has the following limitations and challenges:
The breakpoint of the fusion gene is generally in the intron region, and the intron of the fusion gene is particularly long. Therefore, if the DNA-based detection method is to lay a large number of probes in the intron region, the probe design is difficult.
A large number of probes may also result in excessive data volume on the computer (increasing the experimental cost and the difficulty in analyzing the generated signals).
Covering blind spots still exist for a large number of probes, leading to undetected fusion.
Fusion at the 4.DNA level does not necessarily translate into mRNA, which in turn translates into protein, and does not activate the downstream MAPK/mTOR pathway, whereas RNA is relatively closer to translation into protein.
Compared with DNA level, the fusion gene at RNA level showed the junction between the front and rear exons of the gene, and the fusion point was relatively fixed. This feature provides an inherent advantage for the precise design of probes or primers. Therefore, according to the sequence characteristics of the fusion gene, it is easier to detect the fusion gene at the RNA level than at the DNA level.
 ▲Common detection methods of fusion genes
▲Common detection methods of fusion genes
04. Fusion gene detection: DNA vs RNA
DNA-based NGS
Principle: The probes were designed to cover the regions where fusion might occur, and the target fragments obtained by hybridization capture were sequenced and analyzed.
Through the establishment of the hybridization capture database, mutation, amplification, fusion, tumor mutation load (TMB) and microsatellite instability of a plurality of tumor-related genes can be detected simultaneously by only using DNA, thereby greatly saving the sample size, determining the breakpoint position of gene fusion and the fusion partner at the DNA level and detecting known and unknown gene fusion.
DNA-based NGS sequencing is based on DNA molecules derived from tumor cells. DNA molecules are made up of double-stranded helices that are more stable relative to RNA. Therefore, histological/cytological samples of DNA-based NGS, including fresh tissues, FFPE, hydrothorax, etc., can meet the requirements of detection quality control.
ctDNA can also be extracted by liquid biopsy for sequencing when tissue samples are difficult to obtain or insufficient in size, which RNA-based NGS cannot achieve. However, gene fusion detected by DNA second-generation sequencing is affected by tumor cell content, sample DNA quality, coverage of capture probes, and complex gene variation at DNA level, and may cause missed detection. In addition, with the extensive application of DNA second-generation sequencing in clinical molecular detection, more and more kinase fusions carrying rare partners are found, but some rare fusions cannot produce functional fusion RNA/ protein.
RNA-based NGS
RNA second-generation sequencing allows the detection of fused transcripts at the RNA level by multiplex PCR amplification, anchored multiplex PCR, or hybridization capture banking. Compared with the situation that the second generation sequencing of DNA needs to capture both exon and intron regions with probes, the RNA sequencing only needs to design primers or probes for the exon region, which is relatively simple and economical and is not affected by complex fusion at the DNA level; moreover, the second generation sequencing of RNA can directly and truly reflect the expression of fusion at the transcription level and the types of fusion partner genes.
However, RNA-based NGS testing samples are relatively demanding. In terms of sample quality, fresh tissue was the first choice for RNA-based NGS testing due to the presence of ribonuclease and the fact that RNA was in a single-stranded structure that was very easily degraded. Studies have shown that up to 50% of archived FFPE samples, especially older samples, do not meet the requirements for RNA library building and sequencing and may not pass pre-sequencing quality control. It is currently not recommended to extract RNA from liquid biopsy samples for tumor fusion gene detection, which also limits the application of RNA-based NGS detection.
In a large cohort study of lung adenocarcinoma in the real world by MSKCC and a study of China population, about 10% of target gene fusions or MET jumps are detected in DNA NGS-negative non-small cell lung cancer samples by RNA-based NGS. Compared with DNA level, the fusion gene at RNA level shows the connection between the two gene exons. The relative fixation of fusion points without the influence of introns provides an innate advantage for the accurate design of probes or primers, which is one of the reasons why RNA-based NGS has more advantages than DNA-based NGS in detecting fusion genes.
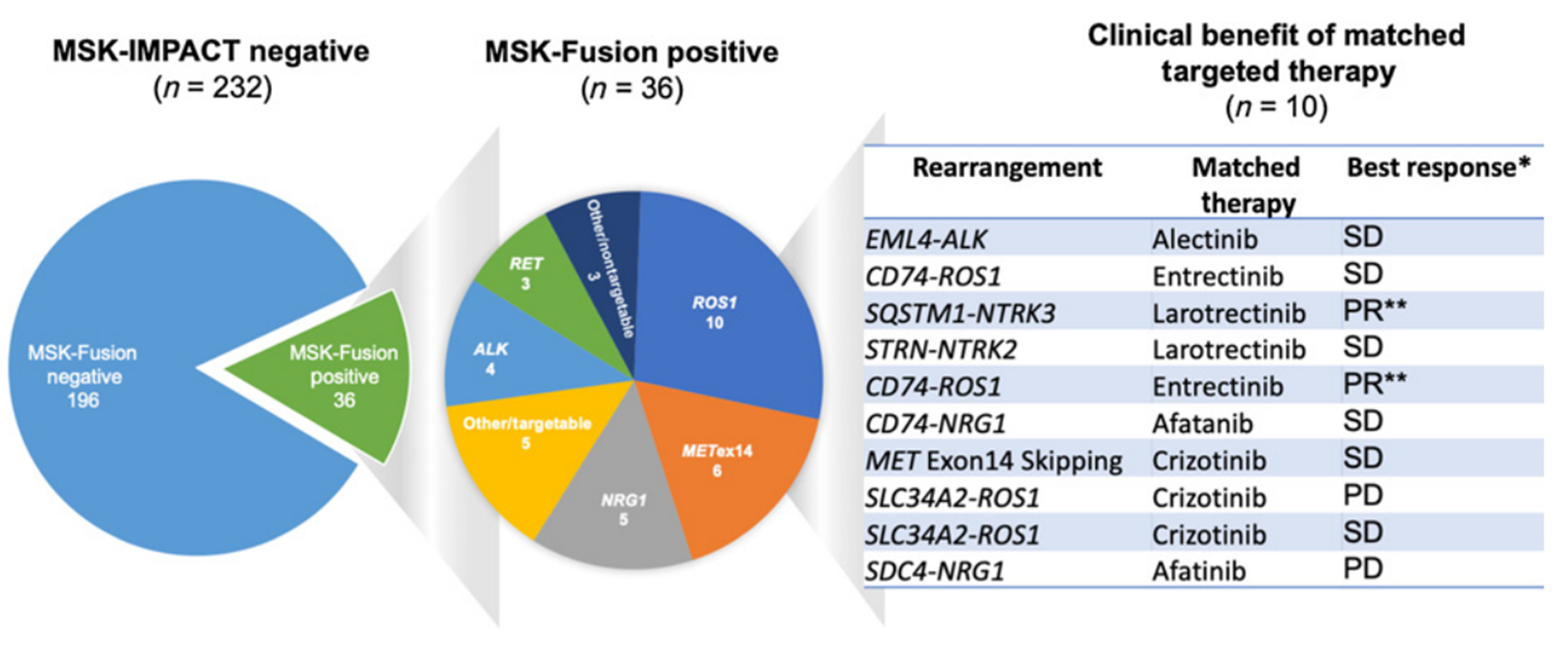
▲Molecular characteristics of fusion genes in lung cancer population from MSKCC
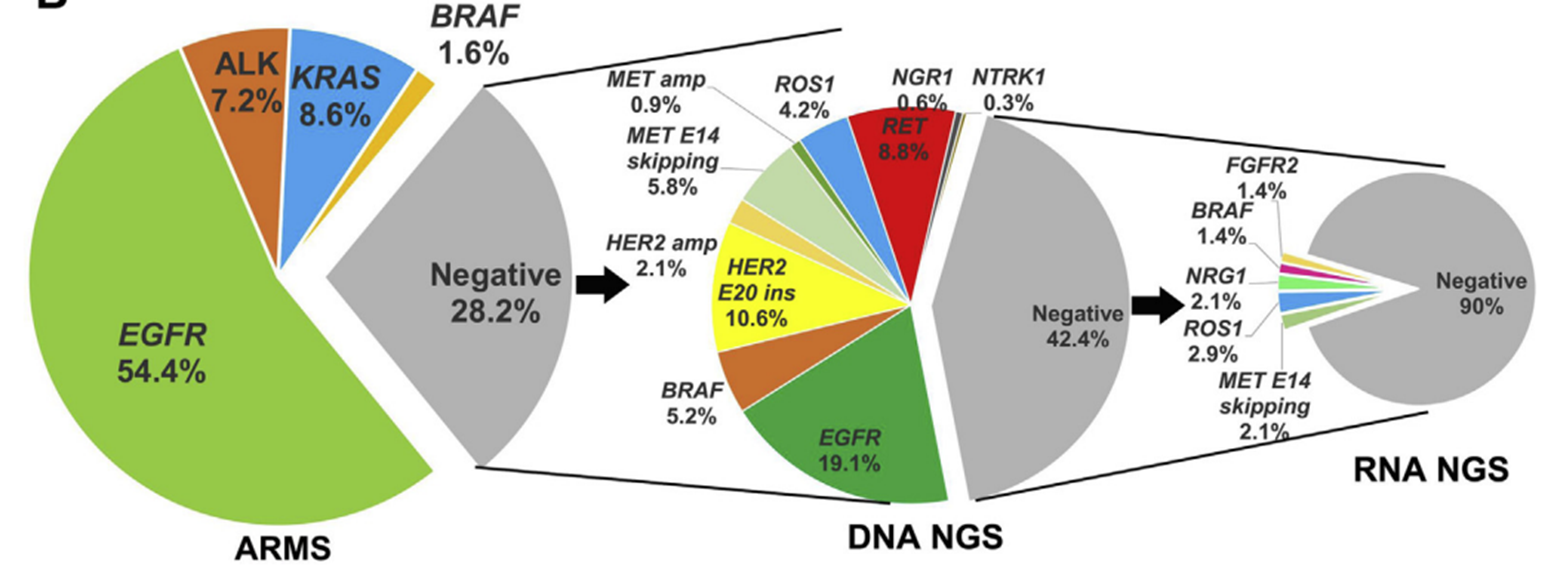
▲ Molecular characteristics of fusion genes in lung cancer population in China
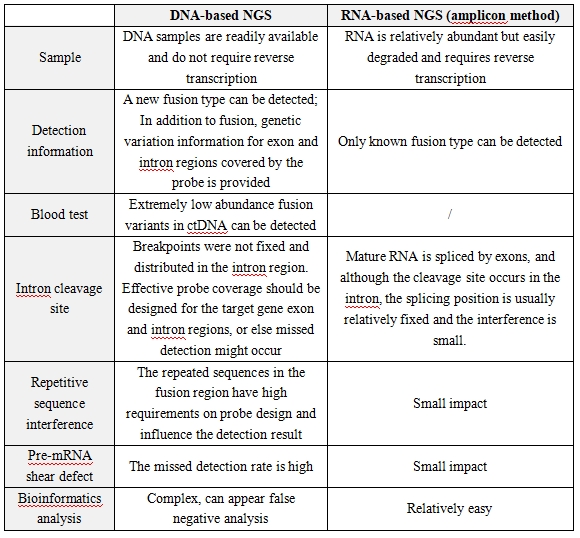
▲ Comparison of NGS detection gene fusion based on DNA and RNA
05. Summary
Compared with DNA-based NGS, RNA-based NGS is not affected by introns, and can improve the detection rate of fusion genes. The structural characteristics at the DNA and RNA levels of fusion genes determine the detection of fusion genes. RNA level is much more sensitive and accurate than DNA level, but the sample requirements are high.
DNA-based NGS have relatively low sample requirements and can be tested in combination with other biomarker, while rare fusion partners, exonic breakpoint fusion and inter-gene fusion can be detected at the DNA level, but further validation at the RNA or protein level is required.
In clinical application, comprehensive and accurate detection of tumor target genes needs the accumulation of clinical practice experience and continuous summary and verification. Due to the limitations of the detection methodology and the complexity of the test specimens, the combined application or supplementation of multiple platforms is beneficial for accurate diagnosis and treatment of tumor patients.
references
[1]Oncotarget. 2019 Mar 12; 10(21):2095-2111.
[2]Genome Biol. 2020 Jul 6; 21(1):166.
[3]Comput Math Methods Med. 2015:2015:912742
[4]DOI:10.3760/cma.j.cn112151-20221111-00946
[5]Noncoding RNA. 2021 Feb 4; 7(1):10
[6]https://doi.org/10.1200/JCO.2024.42.16_suppl.e2004
[7]Clin Cancer Res. 2019 Aug 1; 25(15):4712-4722
[8]J Thorac Oncol. 2021 Mar; 16(3):404-418.
[9]DOI: 10.3779/j.issn.1009-3419.2023.102.43
Statement:
This article is only for sharing, if it involves copyright issues, please contact us as soon as possible, we will correct the first time, thank you!