Current location: Home > NEWS > Industry news
NEWS
PRODUCTS
Prospect of Blood ctDNA in the detection of Glioma
News source: Release time:[2024-07-19]
Circulating tumor DNA(ctDNA) detection has become an important tool for longitudinal assessment of the genomic spectrum in modern clinical oncology with the potential to overcome the limitations of temporal and spatial heterogeneity encountered with tissue sampling. Despite the proven utility of liquid biopsy in many cancers, analysis of ctDNA in gliomas, the most common primary brain malignancy, has not progressed because detection of low levels of circulating glioma DNA remains a challenge.
Cerebrospinal fluid did contain high concentrations of ctDNA, however, since lumbar puncture is not a standard procedure for treatment of glioma patients, it is difficult to obtain duplicate cerebrospinal fluid samples. Other strategies, such as the use of DNA methylene or microRNA in blood or cerebrospinal fluid to analyze epigenetic changes have also shown promise, but these methods lack the specificity of ctDNA to recognize prognostic or targeted mutations in tumors. Blood sampling has a major advantage for liquid biopsies as it can be performed non-invasively at multiple time points. Related exploration aims to evaluate whether the combination of highly sensitive detection method and improved blood sampling method can make plasma liquid biopsy provide valuable genomic information in glioma. In some studies, the first longitudinal study of plasma ctDNA in glioma patients has shown that ctDNA can be successfully detected in the blood of glioma patients, including key genes that can be used to monitor tumor genomic evolution.
Plasma ctDNA enables early detection of temozolomide resistance mutations in gliomas
A total of 10 glioma patients were included in the study, including 8 Grade 4 glioblastoma, 1 Grade 3 IDH1 mutant astrocytoma, and 1 Grade 2 oligodendroglioma. Longitudinal blood sampling was performed before the initial surgery, within 48 hours after surgery, during tumor follow-up (when performing MRI), before the second surgery at relapse, and in some cases after the second surgery. A total of 49 plasma samples (an average of 4 samples per patient) were collected from 10 patients. Collect at least 20 mL of whole blood and at least 5 mL of plasma in an EDTA tube. These samples were tested using the NGS method.
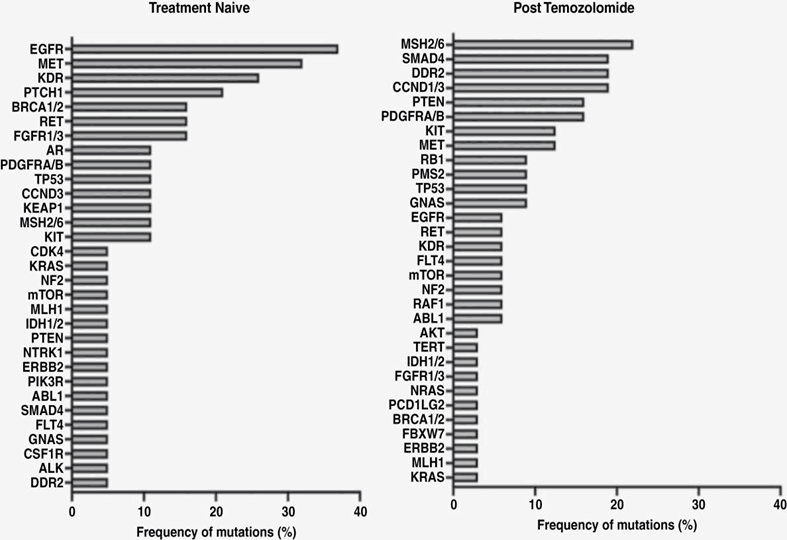
Mutation frequency in patients initially treated with temozolomide and in patients after temozolomide treatment [1]
To evaluate the effect of temozolomide (TMZ) on ctDNA, plasma samples before and after TMZ treatment were compared. Mutations in the MMR gene, including MSH2 or MSH6(20%), PMS(26%), and MLH1, were observed in 7 patients after TMZ treatment. There were no mutations in the MMR gene in PBMC sequencing analyses, suggesting that these MMR mutations found in ctDNA were directly derived from gliomas.
This preliminary study demonstrates that ctDNA based on fluid biopsy is a potential alternative and complementary method for tumor biopsy. In addition, the absence of novel MMR gene mutations in the initial biopsy by plasma ctDNA testing may provide an early indicator of chemoresistance development. Additional clinical verification is required in larger cohorts.
Application of digital PCR blood ctDNA in glioma
A total of 110 glioma patients were included in the study, including 26 Grade 2 gliomas, 13 Grade 3 gliomas, and 71 Grade 4 gliomas. A total of 359 plasma samples were available for ctDNA analysis, with an average of 4 samples per patient.
No significant differences in IDH1m concentration or VAF were observed in 36 patients with mutations in IDH1-R132H. Patients with IDH1m copies/mL and VAF were assigned to high and low groups (medians 0.25 copies/mL and 1.1%, respectively) and no relationship was found between VAF or mutant copies/mL and OS or PFS. No correlation was found between preoperative plasma ID ‑ 1 mVAF or concentration and changes in tumor volume on MRI. Therefore, based on study data, plasma IDH1m concentrations do not appear to be a reliable marker for monitoring tumor size or progression.
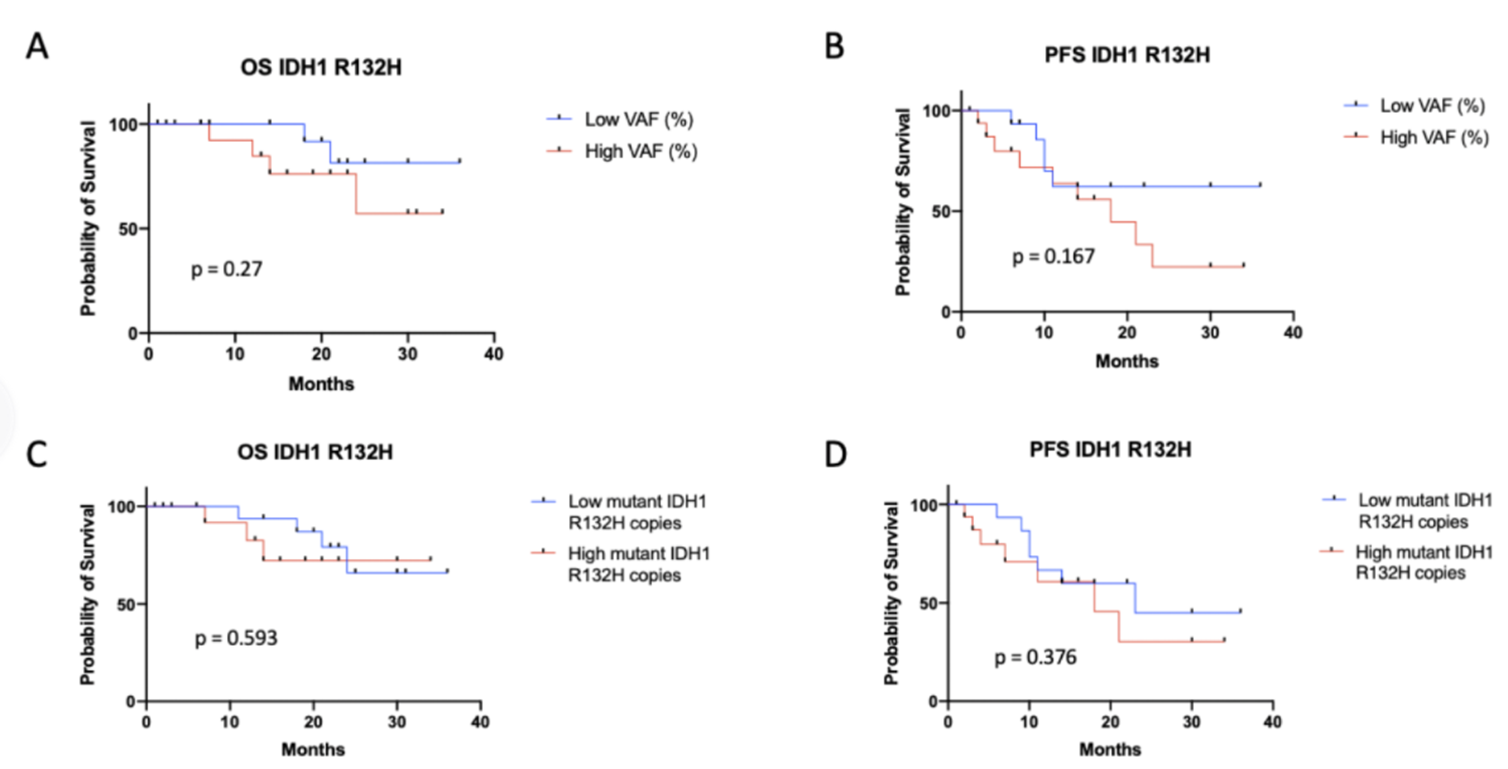 IDH1-R132H Mutation VAF or Mutation copies/mL Not related to OS or PFS [2]
IDH1-R132H Mutation VAF or Mutation copies/mL Not related to OS or PFS [2]
19 patients with TERTp mutation (from tissue NGS detection), 39 of 44 TERTp- positive plasma samples were C228T positive, with a sensitivity of 88.6%. The C250T mutation rate was 48.7%, which was low compared with the detection of NGS mutation in tissues. Neither the concentration of the TERTp mutation nor VAF were significantly associated with survival.
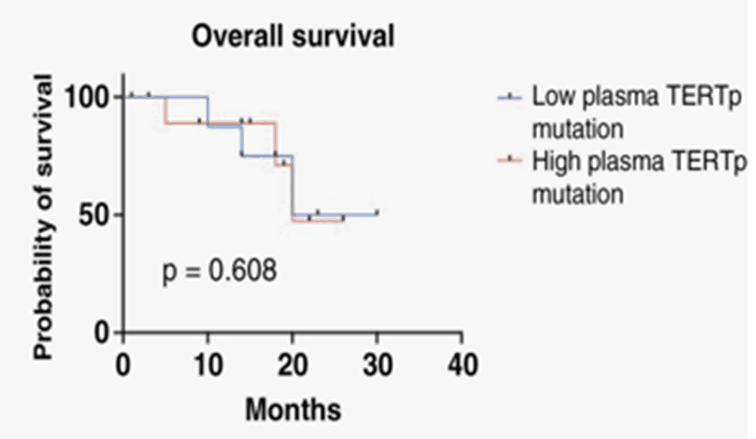
The concentration of TERTp mutations and VAF were not associated with survival [2]
Mutations were detected in plasma in 5 of 7 patients (71%) with organized EGFRvIII, and the overall mean concentrations (0.15 copies /mL) in plasma of EGFRvIII were much lower than those for IDH1-R132H and TERTp mutations. Studies have shown that IDH1-R132H, TERTp, and EGFRvIII can detect mutations in plasma with high sensitivity and specificity, and further studies are needed to verify these findings and the clinical role of ctDNA in gliomas.
Prospect
Mismatch repair defect (MMRd) is caused by acquired mutations of the MMR gene and is another mechanism by which glioma develops TMZ resistance. Even a very small reduction in MSH2/6 activity results in a significant reduction in TMZ susceptibility in vitro. Unlike MGMT methylation, which remains stable during glioma progression, MSH2 and MSH6 are more common in GBMs that recur after TMZ treatment. Performing fluid biopsies to detect MMRd during the treatment and monitoring phases may help to identify TMZ resistance early and begin treatment change to avoid the high risk of mutations associated with persistent TMZ exposure.
Previous literature on ctDNA in gliomas has reported a sensitivity of 50% or less, however, these studies were based on a single plasma sample, whereas in the above studies multiple samples per patient were analyzed. Two additional factors in the study may have contributed to the improved detection rate. First, 2–4 times the volume of plasma reported previously was used and treated rapidly to avoid DNA degradation. Second, NGS testing was optimized for fluid biopsies, increasing sensitivity especially at low DNA concentrations.
At present, the blood ctDNA shows strong specificity in the detection of glioma, and the positive prediction value is high. However, the negative ctDNA result at any time point has weak value in excluding mutations. In the future, more relevant studies are needed to indicate its significance in gliomas and to improve its methods [3].
References
1.Neurooncol Adv. 2024 Mar 19; 6(1):vdae041.
2.Neurooncol Adv. 2024 Mar 4; 6(1):vdae027.
3.Cancer Discov. 2024 Apr 4; 14(4):648-652.
Statement
This article is only for sharing, if it involves copyright issues, please contact us as soon as possible, we will correct the first time, thank you!