Current location: Home > NEWS > Industry news
NEWS
PRODUCTS
As a nucleic acid detection technology, how does digital PCR achieve high sensitivity and absolute quantification?
News source: Release time:[2021-12-06]
Digital PCR (Digital PCR, dPCR) is an absolute nucleicacid quantitative technology that has developed rapidly in recent years after thefirst and second generations of PCR technology.As a brand-new nucleic aciddetection method, digital PCR solves the problem that qPCR needs to rely on astandard curve for quantitative analysis.
By the end-point detection method, the number oftarget DNA molecules can be directly counted, and the absolute quantificationof "single-molecule template amplification" can be realized, whichgreatly improves the detection sensitivity. Digital PCR is currently the mostsensitive nucleic acid detection method. How does it achieve high sensitivityand absolute quantification?
01
One of the biggest differences between digital PCR andfluorescence quantitative PCR is the redistribution of the reaction system.Distribute a sample equally to tens of thousands of reaction units, so that theDNA in each reaction unit is amplified separately. Use fluorescent probes ordye methods to detect specific target sequences, and statistically analyze thefluorescent signals of each reaction unit after the amplification is completed.
This article starts with theadvantages of partitioning and Poisson statistics, including the sources oferrors, discussing the principles of digital PCR.
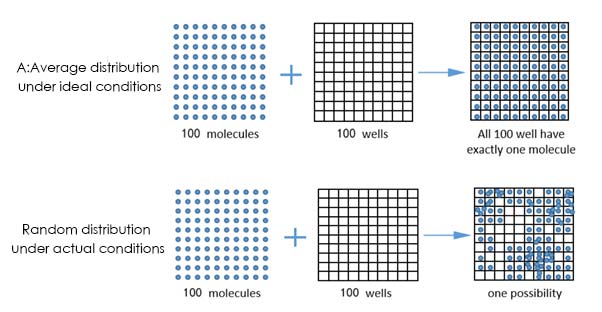
The first step is the limited dilution. In order toachieve single-molecule template amplification, the diluted PCR reaction systemcontain the target molecule is divided into tens of thousands of tiny reactionunits, so that each tiny unit contains a discrete number of target genes (0, 1,2, 3,...).
That is to say, dozens of microliters of PCR reactionsystem that contains primers, templates, buffers, enzymes and other componentsare divided into many tiny independent reaction systems, so that the number ofDNA molecules can be directly counted. The initial sample is absolutelyquantified, by dividing a sample into tens of thousands to millions of partsand assigning them to different reaction units, each unit contains one or moretarget genes.
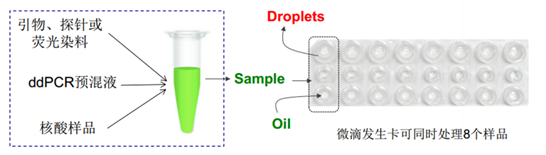
Droplet preparation: 20μL system can generate 20,000droplets. Conversion: 1 liter = 1000 ml = 1,000,000 microliters, 1 microliter =0.001 ml = 0.000001 liters.

The second step is the amplification reaction. Eachtiny unit undergoes independent PCR amplification, the fluorescent signal ofeach reaction unit is detected after the amplification is completed.
Tens of thousands of microsystems after PCRamplification are subjected to fluorescence detection. Statistically speaking, Ifthe system containing the target gene is amplified and the fluorescent signalcan be detected, it is marked as 1 (positive). If the fluorescent signal cannotbe detected in the system that does not contain the template, it is marked as 0(negative). However, since digital PCR is a terminal analysis method for theresults after the amplification, for example, the distribution of the targetgene in the reagent operation is not uniform, but discretized.
Even some small droplets have more than one target atthe end (for example, there are several mutant targets in one droplet at thesame time, such as KRAS, EGFR, BRAF, and TP53), then theoretically theresulting concentration may be discarded.
The third step is to statistically analyze the numberand proportion of negative reactions according to the Poisson distributionprinciple, and directly calculate the concentration or copy number of thetested target molecule.
It is known that many statistical results conform to anormal distribution. That is, a symmetrical distribution with a small number atboth ends and a large number at the middle.
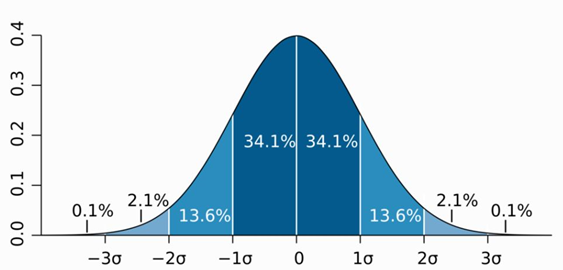
According to the principle of normal distribution, thetotal number and the highest sub-peak value are known, therefore various datain other sections can often be calculated.
▲Poisson Distribution Chart
The principle is similar for digital PCR. For example,if we divide a small PCR system into 20,000 copies, we know the total number.At this time, we only need to count the number of extreme values, and then wecan accurately calculate the data we need for other sections according to theprinciple of Poisson distribution.
02
So why does our digital PCR choose Poissondistribution statistics instead of the normal distribution statistical model?What is the difference between these two models? In fact, when we distributePCR droplets, a limited number of distributions (20,000 droplets) are carriedout over a period of time. The statistical formula is more in line with thePoisson distribution. If the time axis continues to be extended to a year oreven a few years, the sample was distributed into trillions of droplets.
Therefore, in terms of practical operability, thestatistics in real work use the Poisson distribution model more practically.
Spacegen
Spacegen focuses on the development of precision tumormedicine, based on digital PCR technology, develops related products, and leadsinnovation.
NO. | Product Name | Detection Item | Matched Instrument |
1 | Human EGFR Gene T790M Mutation Detection Kit | EGFR-T790M | Bio-Rad QX100/200 |
2 | Human EGFR Gene L858R Mutation Detection Kit | EGFR-L858R | |
3 | Human EGFR Gene 19-del Mutation Detection Kit | EGFR-19-del | |
5 | Human HER2 Gene Amplification Detection Kit | Her2 gene amplification | |
6 | Human MET Gene Amplification Detection Kit | MET gene amplification |